Abstract
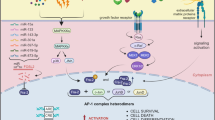
Among the FOS-related members of the AP-1 dimeric complex, the transcription factor Fra-1, encoded by FOSL1, is crucially involved in human tumor progression and metastasis, thus representing a promising therapeutic target. Here we review the state of the art and discuss the emerging topics and perspectives on FOSL1 and its gene product. First, we summarize the present knowledge on the FOSL1 transcriptional and epigenetic controls, driving Fra-1 accumulation in a variety of human solid tumors. We also present a model on the regulatory interactions between Fra-1, p53, and miRNAs. Then, we outline the multiple roles of Fra-1 posttranslational modifications and transactivation mechanisms of select Fra-1 target genes. In addition to summarizing the Fra-1-dependent gene networks controlling proliferation, survival, and epithelial–mesenchymal transitions (EMT) in multiple cancer cell types, we highlight the roles played by Fra-1 in nonneoplastic cell populations recruited to the tumor microenvironment, and in mouse models of tumorigenesis. Next, we review the prognostic power of the Fra-1-associated gene signatures, and envisage potential strategies aimed at Fra-1 therapeutic inhibition. Finally, we discuss several recent reports showing the emerging roles of Fra-1 in the mechanisms of both resistance and addiction to targeted therapies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3:859–68.
Jiang X, Xie H, Dou Y, Yuan J, Zeng D, Xiao S. Expression and function of FRA1 protein in tumors. Mol Biol Rep. 2020;47:737–52.
Dhillon AS, Tulchinsky E. FRA-1 as a driver of tumour heterogeneity: a nexus between oncogenes and embryonic signalling pathways in cancer. Oncogene. 2015;34:4421–8.
Casalino L, De Cesare D, Verde P. Accumulation of Fra-1 in ras-transformed cells depends on both transcriptional autoregulation and MEK-dependent posttranslational stabilization. Mol Cell Biol. 2003;23:4401–15.
Liu W, Tian T, Liu L, Du J, Gu Y, Qin N, et al. A functional SNP rs1892901 in FOSL1 is associated with gastric cancer in Chinese population. Sci Rep. 2017;7:41737.
Zhang L, Liu H, Mu X, Cui J, Peng Z. Dysregulation of Fra1 expression by Wnt/β-catenin signalling promotes glioma aggressiveness through epithelial–mesenchymal transition. Biosci Rep. 2017;37:BSR20160643.
Liu H, Ren G, Wang T, Chen Y, Gong C, Bai Y, et al. Aberrantly expressed Fra-1 by IL-6/STAT3 transactivation promotes colorectal cancer aggressiveness through epithelial-mesenchymal transition. Carcinogenesis. 2015;36:459–68.
Zippo A, Serafini R, Rocchigiani M, Pennacchini S, Krepelova A, Oliviero S. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell. 2009;138:1122–36.
Esnault C, Gualdrini F, Horswell S, Kelly G, Stewart A, East P, et al. ERK-induced activation of TCF family of SRF cofactors initiates a chromatin modification cascade associated with transcription. Mol Cell. 2017;65:1081–.e5.
Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–34.
Lockwood WW, Zejnullahu K, Bradner JE, Varmus H. Sensitivity of human lung adenocarcinoma cell lines to targeted inhibition of BET epigenetic signaling proteins. Proc Natl Acad Sci USA. 2012;109:19408–13.
Baker EK, Taylor S, Gupte A, Sharp PP, Walia M, Walsh NC, et al. BET inhibitors induce apoptosis through a MYC independent mechanism and synergise with CDK inhibitors to kill osteosarcoma cells. Sci Rep. 2017;5:10120.
Zhao C, Qiao Y, Jonsson P, Wang J, Xu L, Rouhi P, et al. Genome-wide profiling of AP-1-regulated transcription provides insights into the invasiveness of triple-negative breast cancer. Cancer Res. 2014;74:3983–94.
Zanconato F, Forcato M, Battilana G, Azzolin L, Quaranta E, Bodega B, et al. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat Cell Biol. 2015;17:1218–27.
Zanconato F, Battilana G, Forcato M, Filippi L, Azzolin L, Manfrin A, et al. Transcriptional addiction in cancer cells is mediated by YAP/TAZ through BRD4. Nat Med. 2018;24:1599–610.
Tam WL, Lu H, Buikhuisen J, Soh BS, Lim E, Reinhardt F, et al. Protein kinase C α is a central signaling node and therapeutic target for breast cancer stem cells. Cancer Cell. 2013;24:347–64.
Lin Y, Yao J, Zeng Y-X. Disrupting the interaction of BRD4 with diacetylated twist suppresses tumorigenesis in basal-like breast cancer. Cancer Cell. 2014;25:210–25.
Desmet CJ, Gallenne T, Prieur A, Reyal F, Visser NL, Wittner BS, et al. Identification of a pharmacologically tractable Fra-1/ADORA2B axis promoting breast cancer metastasis. Proc Natl Acad Sci USA. 2013;110:5139–44.
Ding X, Pan H, Li J, Zhong Q, Chen X, Dry SM, et al. Epigenetic activation of AP1 promotes squamous cell carcinoma metastasis. Sci Signal. 2013;6:ra28.1–13. – S0–15
Amundson SA, Bittner M, Chen Y, Trent J, Meltzer P, Fornace AJ. Fluorescent cDNA microarray hybridization reveals complexity and heterogeneity of cellular genotoxic stress responses. Oncogene. 1999;18:3666–72.
Nikulenkov F, Spinnler C, Li H, Tonelli C, Shi Y, Turunen M, et al. Insights into p53 transcriptional function via genome-wide chromatin occupancy and gene expression analysis. Cell Death Differ. 2012;19:1992–2002.
Li M, He Y, Dubois W, Wu X, Shi J, Huang J. Distinct regulatory mechanisms and functionsfor p53-activated and p53-repressed DNA damage response genes in embryonic stem cells. Mol Cell. 2012;46:30–42.
Wu J, Wu G, Lv L, Ren Y-F, Zhang X-J, Xue Y-F, et al. MicroRNA-34a inhibits migration and invasion of colon cancer cells via targeting to Fra-1. Carcinogenesis. 2012;33:519–28.
Yang S, Li Y, Gao J, Zhang T, Li S, Luo A, et al. MicroRNA-34 suppresses breast cancer invasion and metastasis by directly targeting Fra-1. Oncogene. 2013;32:4294–303.
Zhang N, Shen Q, Zhang P. miR-497 suppresses epithelial-mesenchymal transition and metastasis in colorectal cancer cells by targeting fos-related antigen-1. Onco Targets Ther. 2016;9:6597–604.
Wu J, Ji A, Wang X, Zhu Y, Yu Y, Lin Y, et al. MicroRNA-195-5p, a new regulator of Fra-1, suppresses the migration and invasion of prostate cancer cells. J Transl Med. 2015;13:289.
Jin Y, Wang C, Liu X, Mu W, Chen Z, Yu D, et al. Molecular characterization of the MicroRNA-138-Fos-like antigen 1 (FOSL1) regulatory module in squamous cell carcinoma. J Biol Chem. 2011;286:40104–9.
Rattanasinchai C, Llewellyn BJ, Conrad SE, Gallo KA. MLK3 regulates FRA-1 and MMPs to drive invasion and transendothelial migration in triple-negative breast cancer cells. Oncogenesis. 2017;6:e345.
Belguise K, Cherradi S, Sarr A, Boissière F, Boulle N, Simony-Lafontaine J, et al. PKCθ-induced phosphorylations control the ability of Fra-1 to stimulate gene expression and cancer cell migration. Cancer Lett. 2017;385:97–107.
Wang T, Song P, Zhong T, Wang X, Xiang X, Liu Q, et al. The inflammatory cytokine IL-6 induces FRA1 deacetylation promoting colorectal cancer stem-like properties. Oncogene. 2019;38:4932–47.
Liu X, Li H, Rajurkar M, Li Q, Cotton JL, Ou J, et al. Tead and AP1 coordinate transcription and motility. Cell Rep. 2016;14:1169–80.
Fleming JD, Pavesi G, Benatti P, Imbriano C, Mantovani R, Struhl K. NF-Y coassociates with FOS at promoters, enhancers, repetitive elements, and inactive chromatin regions, and is stereo-positioned with growth-controlling transcription factors. Genome Res. 2013;23:1195–209.
Bejjani F, Evanno E, Zibara K, Piechaczyk M, Jariel-Encontre I. The AP-1 transcriptional complex: local switch or remote command? BBA Rev Cancer. 2019;1872:11–23.
Nerlov C, De Cesare D, Pergola F, Caracciolo A, Blasi F, Johnsen M, et al. A regulatory element that mediates co-operation between a PEA3-AP-1 element and an AP-1 site is required for phorbol ester induction of urokinase enhancer activity in HepG2 hepatoma cells. EMBO J. 1992;11:4573–82.
Natoli G, Andrau J-C. Noncoding transcription at enhancers: general principles and functional models. Annu Rev Genet. 2012;46:1–19.
Moquet-Torcy G, Tolza C, Piechaczyk M, Jariel-Encontre I. Transcriptional complexity and roles of Fra-1/AP-1 at the uPA/Plau locus in aggressive breast cancer. Nucleic Acids Res. 2014;42:11011–24.
Franco HL, Nagari A, Malladi VS, Li W, Xi Y, Richardson D, et al. Enhancer transcription reveals subtype-specific gene expression programs controlling breast cancer pathogenesis. Genome Res. 2018;28:159–70.
Tolza C, Bejjani F, Evanno E, Mahfoud S, Moquet-Torcy G, Gostan T, et al. AP-1 signaling by Fra-1 directly regulates HMGA1 oncogene transcription in triple-negative breast cancers. Mol Cancer Res. 2019;17:1999–2014.
Qiao Y, Shiue C-N, Zhu J, Zhuang T, Jonsson P, Wright APH, et al. AP-1-mediated chromatin looping regulates ZEB2 transcription: new insights into TNFα-induced epithelial–mesenchymal transition in triple-negative breast cancer. Oncotarget. 2015;6:7804–14.
Evellin S, Galvagni F, Zippo A, Neri F, Orlandini M, Incarnato D, et al. FOSL1 controls the assembly of endothelial cells into capillary tubes by direct repression of αv and β3 integrin transcription. Mol Cell Biol. 2013;33:1198–209.
He H, Song D, Sinha I, Hessling B, Li X, Haldosen L-A, et al. Endogenous interaction profiling identifies DDX5 as an oncogenic coactivator of transcription factor Fra-1. Oncogene. 2019;38:5725–38.
Shin S, Rossow KL, Grande JP, Janknecht R. Involvement of RNA helicases p68 and p72 in colon cancer. Cancer Res. 2007;67:7572–8.
Yang L, Lin C, Liu Z-R. P68 RNA helicase mediates PDGF-induced epithelial mesenchymal transition by displacing Axin from β-catenin. Cell. 2006;127:139–55.
Dardenne E, Polay Espinoza M, Fattet L, Germann S, Lambert M-P, Neil H, et al. RNA helicases DDX5 and DDX17 dynamically orchestrate transcription, miRNA, and splicing programs in cell differentiation. Cell Rep. 2014;7:1900–13.
Racca AC, Prucca CG, Caputto BL. Fra-1 and c-Fos N-terminal deletion mutants impair breast tumor cell proliferation by blocking lipid synthesis activation. Front Oncol. 2019;9:544.
Blunsom NJ, Cockcroft S. CDP-diacylglycerol synthases (CDS): gateway to phosphatidylinositol and cardiolipin synthesis. Front Cell Dev Biol. 2020;8:63.
Zhang X, Wu J, Luo S, Lechler T, Zhang JY. FRA1 promotes squamous cell carcinoma growth and metastasis through distinct AKT and c-Jun dependent mechanisms. Oncotarget. 2016;7:34371–83.
Casalino L, Bakiri L, Talotta F, Weitzman JB, Fusco A, Yaniv M, et al. Fra-1 promotes growth and survival in RAS-transformed thyroid cells by controlling cyclin A transcription. EMBO J. 2007;26:1878–90.
Sayan AE, Stanford R, Vickery R, Grigorenko E, Diesch J, Kulbicki K, et al. Fra-1 controls motility of bladder cancer cells via transcriptional upregulation of the receptor tyrosine kinase AXL. Oncogene. 2012;31:1493–503.
Vallejo A, Perurena N, Guruceaga E, Mazur PK, Martinez-Canarias S, Zandueta C, et al. An integrative approach unveils FOSL1 as an oncogene vulnerability in KRAS-driven lung and pancreatic cancer. Nat Commun. 2017;8:14294–14.
Belguise K, Kersual N, Galtier F, Chalbos D. FRA-1 expression level regulates proliferation and invasiveness of breast cancer cells. Oncogene. 2005;24:1434–44.
Bakiri L, Macho-Maschler S, Custic I, Niemiec J, Guío-Carrión A, Hasenfuss SC, et al. Fra-1/AP-1 induces EMT in mammary epithelial cells by modulating Zeb1/2 and TGFβ expression. Cell Death Differ. 2014;22:336–50.
Walia V, Ding M, Kumar S, Nie D, Premkumar LS, Elble RC. hCLCA2 Is a p53-inducible inhibitor of breast cancer cell proliferation. Cancer Res. 2009;69:6624–32.
Vial E, Sahai E, Marshall CJ. ERK-MAPK signaling coordinately regulates activity of Rac1 and RhoA for tumor cell motility. Cancer Cell. 2003;4:67–79.
Iskit S, Schlicker A, Wessels L, Peeper DS. Fra-1 is a key driver of colon cancer metastasis and a Fra-1 classifier predicts disease-free survival. Oncotarget. 2015;5:43146–61.
Ibrahim SAE-F, Abudu A, Jonhson E, Aftab N, Conrad S, Fluck M. The role of AP-1 in self-sufficient proliferation and migration of cancer cells and its potential impact on an autocrine/paracrine loop. Oncotarget. 2018;9:34259–78.
Elangovan IM, Vaz M, Tamatam CR, Potteti HR, Reddy NM, Reddy SP. FOSL1 promotes Kras-induced lung cancer through amphiregulin and cell survival gene regulation. Am J Respir Cell Mol Biol. 2017;58:625–35.
He J, Zhu G, Gao L, Chen P, Long Y, Liao S, et al. Fra-1 is upregulated in gastric cancer tissues and affects the PI3K/Akt and p53 signaling pathway in gastric cancer. Int J Oncol. 2015;47:1725–34.
Zhong G, Chen X, Fang X, Wang D, Xie M, Chen Q. Fra-1 is upregulated in lung cancer tissues and inhibits the apoptosis of lung cancer cells by the P53 signaling pathway. Oncol Rep. 2015;35:447–53.
Xiao S, Zhou Y, Yi W, Luo G, Jiang B, Tian Q, et al. Fra‑1 is downregulated in cervical cancer tissues and promotes cervical cancer cell apoptosis by p53 signaling pathway in vitro. Int J Oncol. 2015;46:1677–84.
Walia V, Yu Y, Cao D, Sun M, McLean JR, Hollier BG, et al. Loss of breast epithelial marker hCLCA2 promotes epithelial-to-mesenchymal transition and indicates higher risk of metastasis. Oncogene. 2012;31:2237–46.
Andreolas C, Kalogeropoulou M, Voulgari A, Pintzas A. Fra-1 regulates vimentin during Ha-RAS-induced epithelial mesenchymal transition in human colon carcinoma cells. Int J Cancer. 2008;122:1745–56.
Diesch J, Sanij E, Gilan O, Love C, Tran H, Fleming NI, et al. Widespread FRA1-dependent control of mesenchymal transdifferentiation programs in colorectal cancer cells. PLoS ONE. 2014;9:e88950.
Sundqvist A, Zieba A, Vasilaki E, Herrera Hidalgo C, Söderberg O, Koinuma D, et al. Specific interactions between Smad proteins and AP-1 components determine TGFβ-induced breast cancer cell invasion. Oncogene. 2012;32:3606–15.
Cheng F, Su L, Yao C, Liu L, Shen J, Liu C, et al. SIRT1 promotes epithelial-mesenchymal transition and metastasis in colorectal cancer by regulating fra-1 expression. Cancer Lett. 2016. https://doi.org/10.1016/j.canlet.2016.03.010.
Zhang K, Myllym kiS-M, Gao P, Devarajan R, Kyt lV, Nykter M, et al. Oncogenic K-Ras upregulates ITGA6 expression via FOSL1 to induce anoikis resistance and synergizes with aV-Class integrins to promote EMT. Oncogene. 2017;36:5681–94.
Caramel J, Papadogeorgakis E, Hill L, Browne GJ, Richard G, Wierinckx A, et al. A switch in the expression of embryonic EMT-inducers drives the development of malignant melanoma. Cancer Cell. 2013;24:466–80.
Maurus K, Hufnagel A, Geiger F, Graf S, Berking C, Heinemann A, et al. The AP-1 transcription factor FOSL1 causes melanocyte reprogramming and transformation. Oncogene. 2017;36:5110–21.
Tulchinsky E, Pringle JH, Caramel J, Ansieau S. Plasticity of melanoma and EMT-TF reprogramming. Oncotarget. 2014;5:1–2.
Toyozumi T, Hoshino I, Takahashi M, Usui A, Akutsu Y, Hanari N, et al. Fra-1 regulates the expression of HMGA1, which is associated with a poor prognosis in human esophageal squamous cell carcinoma. Ann Surg Oncol. 2016;24:3446–55.
Dikshit A, Jin YJ, Degan S, Hwang J, Foster MW, Li C-Y, et al. UBE2N promotes melanoma growth via MEK/FRA1/SOX10 signaling. Cancer Res. 2018;78:6462–72.
Tulchinsky E, Demidov O, Kriajevska M, Barlev NA, Imyanitov E. EMT: a mechanism for escape from EGFR-targeted therapy in lung cancer. BBA Rev Cancer. 2019;1871:29–39.
Stinson S, Lackner MR, Adai AT, Yu N, Kim H-J, O’Brien C, et al. TRPS1 targeting by miR-221/222 promotes the epithelial-to-mesenchymal transition in breast cancer. Sci Signal. 2011;4:ra41.
Wu J, Sun Y, Zhang P-Y, Qian M, Zhang H, Chen X, et al. The Fra-1–miR-134–SDS22 feedback loop amplifies ERK/JNK signaling and reduces chemosensitivity in ovarian cancer cells. Cell Death Dis. 2016;7:e2384.
Iaccarino I. lncRNAs and MYC: an intricate relationship. IJMS. 2017;18:1497.
Léveillé N, Melo CA, Rooijers K, Díaz-Lagares A, Melo SA, Korkmaz G, et al. Genome-wide profiling of p53-regulated enhancer RNAs uncovers a subset of enhancers controlled by a lncRNA. Nat Commun. 2015;6:6520.
Luo YP, Zhou H, Krueger J, Kaplan C, Liao D, Markowitz D, et al. The role of proto-oncogene Fra-1 in remodeling the tumor microenvironment in support of breast tumor cell invasion and progression. Oncogene. 2010;29:662–73.
Wang Q, Ni H, Lan L, Wei X, Xiang R, Wang Y. Fra-1 protooncogene regulates IL-6 expression in macrophages and promotes the generation of M2d macrophages. Cell Res. 2010;20:701–12.
Yang J, Zhang Z, Chen C, Liu Y, Si Q, Chuang TH, et al. MicroRNA-19a-3p inhibits breast cancer progression and metastasis by inducing macrophage polarization through downregulated expression of Fra-1 proto-oncogene. Oncogene. 2014;33:3014–23.
Lau EYT, Lo J, Cheng BYL, Ma MKF, Lee JMF, Ng JKY, et al. Cancer-associated fibroblasts regulate tumor-initiating cell plasticity in hepatocellular carcinoma through c-Met/FRA1/HEY1 signaling. Cell Rep. 2016;15:1175–89.
Ramos-Nino ME, Scapoli L, Martinelli M, Land S, Mossman BT. Microarray analysis and RNA silencing link fra-1 to cd44 and c-met expression in mesothelioma. Cancer Res. 2003;63:3539–45.
Ramos-Nino ME, Blumen SR, Sabo-Attwood T, Pass H, Carbone M, Testa JR, et al. HGF mediates cell proliferation of human mesothelioma cells through a PI3K/MEK5/Fra-1 pathway. Am J Respir Cell Mol Biol. 2008;38:209–17.
Gallenne T, Ross KN, Visser NL, Salony. S, Desmet CJ, Wittner BS, et al. Systematic functional perturbations uncover a prognostic genetic network driving human breast cancer. Oncotarget. 2017;8:20572–87.
Román M, López I, Guruceaga E, Baraibar I, Ecay M, Collantes M, et al. Inhibitor of differentiation-1 sustains mutant KRAS-driven progression, maintenance, and metastasis of lung adenocarcinoma via regulation of a FOSL1 network. Cancer Res. 2019;79:625–38.
Behan FM, Iorio F, Picco G, Gonçalves E, Beaver CM, Migliardi G, et al. Prioritization of cancer therapeutic targets using CRISPR–Cas9 screens. Nature. 2019;568:511–6.
Luo Y, Zhou H, Mizutani M, Mizutani N, Reisfeld RA, Xiang R. Transcription factor Fos-related antigen 1 is an effective target for a breast cancer vaccine. Proc Natl Acad Sci USA. 2003;100:8850–5.
Chandran SS, Klebanoff CA. T cell receptor‐based cancer immunotherapy: emerging efficacy and pathways of resistance. Immunol Rev. 2019;290:127–47.
Kamide D, Yamashita T, Araki K, Tomifuji M, Tanaka Y, Tanaka S, et al. Selective activator protein‐1 inhibitor T‐5224 prevents lymph node metastasis in an oral cancer model. Cancer Sci. 2016;107:666–73.
Blake DR, Vaseva AV, Hodge RG, Kline MP, Gilbert TSK, Tyagi V, et al. Application of a MYC degradation screen identifies sensitivity to CDK9 inhibitors in KRAS-mutant pancreatic cancer. Sci Signal. 2019;12:eaav7259.
Zhou J, Rossi J. Aptamers as targeted therapeutics: current potential and challenges. Nat Rev Drug Discov. 2016;16:181–202.
Kroger C, Afeyan A, Mraz J, Eaton EN, Reinhardt F, Khodor YL, et al. Acquisition of a hybrid E/M state is essential for tumorigenicity of basal breast cancer cells. PNAS. 2019;8:201812876–7362.
Ishay-Ronen D, Diepenbruck M, Kalathur RKR, Sugiyama N, Tiede S, Ivanek R, et al. Gain fat—lose metastasis: converting invasive breast cancer cells into adipocytes inhibits cancer metastasis. Cancer Cell. 2019;35:17–32.e6.
Gerdes MJ, Myakishev M, Frost NA, Rishi V, Moitra J, Acharya A, et al. Activator protein-1 activity regulates epithelial tumor cell identity. Cancer Res. 2006;66:7578–88.
Luther J, Driessler F, Megges M, Hess A, Herbort B, Mandic V, et al. Elevated Fra-1 expression causes severe lipodystrophy. J Cell Sci. 2011;124:1465–76.
Hasenfuss SC, Bakiri L, Thomsen MK, Williams EG, Auwerx J, Wagner EF. Regulation of steatohepatitis and PPARγ signaling by distinct AP-1 dimers. Cell Metab. 2014;19:84–95.
Obenauf AC, Zou Y, Ji AL, Vanharanta S, Shu W, Shi H, et al. Therapy-induced tumour secretomes promote resistance and tumour progression. Nature. 2015;520:368–72.
Grimm J, Hufnagel A, Wobser M, Borst A, Haferkamp S, Houben R, et al. BRAF inhibition causes resilience of melanoma cell lines by inducing the secretion of FGF1. Oncogenesis. 2018;7:71.
Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–4.
Duncan JS, Whittle MC, Nakamura K, Abell AN, Midland AA, Zawistowski JS, et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell. 2012;149:307–21.
Kurimchak AM, Shelton C, Herrera-Montávez C, Duncan KE, Chernoff J, Duncan JS. Intrinsic resistance to MEK inhibition through BET protein-mediated kinome reprogramming in NF1-deficient ovarian cancer. Mol Cancer Res. 2019;17:1721–34.
Shin S, Dimitri CA, Yoon S-O, Dowdle W, Blenis J. ERK2 but not ERK1 induces epithelial-to-mesenchymal transformation via DEF motif-dependent signaling events. Mol Cell. 2010;38:114–27.
Kong X, Kuilman T, Shahrabi A, Boshuizen J, Kemper K, Song J-Y, et al. Cancer drug addiction is relayed by an ERK2-dependent phenotype switch. Nature. 2017;550:270–4.
Hong A, Moriceau G, Sun L, Lomeli S, Piva M, Damoiseaux R, et al. Exploiting drug addiction mechanisms to select against MAPKi-resistant melanoma. Cancer Discov. 2018;8:74–93.
Dongre A, Rashidian M, Reinhardt F, Bagnato A, Keckesova Z, Ploegh HL, et al. Epithelial-to-mesenchymal transition contributes to immunosuppression in breast carcinomas. Cancer Res. 2017;77:3982–9.
Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165:35–44.
Lee M-H, Yanagawa J, Tran L, Walser TC, Bisht B, Fung E, et al. FRA1 contributes to MEK-ERK pathway-dependent PD-L1 upregulation by KRAS mutation in premalignant human bronchial epithelial cells. Am J Transl Res. 2020;12:409–27.
Acknowledgements
We wish to thank all the authors whose work was not cited here owing to page limitation. We thank the former members of the lab for fruitful discussions, along with Valerio Costa and Ingram Iaccarino for critical feedback on the paper. We also thank the Regione Campania Project POR-SATIN for financial support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Talotta, F., Casalino, L. & Verde, P. The nuclear oncoprotein Fra-1: a transcription factor knocking on therapeutic applications’ door. Oncogene 39, 4491–4506 (2020). https://doi.org/10.1038/s41388-020-1306-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-020-1306-4
This article is cited by
-
EMT/MET plasticity in cancer and Go-or-Grow decisions in quiescence: the two sides of the same coin?
Molecular Cancer (2023)
-
Multiple Fra-1-bound enhancers showing different molecular and functional features can cooperate to repress gene transcription
Cell & Bioscience (2023)
-
The identification of genes associated T-cell exhaustion and construction of prognostic signature to predict immunotherapy response in lung adenocarcinoma
Scientific Reports (2023)
-
Loss of function of GATA3 regulates FRA1 and c-FOS to activate EMT and promote mammary tumorigenesis and metastasis
Cell Death & Disease (2023)
-
Maternal Undernutrition Induces Cell Signalling and Metabolic Dysfunction in Undifferentiated Mouse Embryonic Stem Cells
Stem Cell Reviews and Reports (2023)