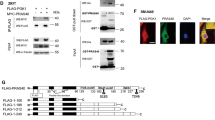
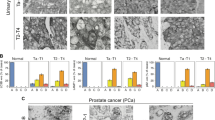
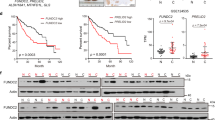
Abstract
Mutants in the gene encoding mitochondrion-associated protein LRPPRC were found to be associated with French Canadian Type Leigh syndrome, a human disorder characterized with neurodegeneration and cytochrome c oxidase deficiency. LRPPRC interacts with one of microtubule-associated protein family MAP1S that promotes autophagy initiation and maturation to suppress genomic instability and tumorigenesis. Previously, although various studies have attributed LRPPRC nuclear acid-associated functions, we characterized that LRPPRC acted as an inhibitor of autophagy in human cancer cells. Here we show that liver-specific deletion of LRPPRC causes liver-specific increases of YAP and P27 and decreases of P62, leading to an increase of cell polyploidy and an impairment of autophagy maturation. The blockade of autophagy maturation and promotion of polyploidy caused by LRPPRC depletion synergistically enhances diethylnitrosamine-induced DNA damage, genome instability, and further tumorigenesis so that LRPPRC knockout mice develop more and larger hepatocellular carcinomas and survive a shorter lifespan. Therefore, LRPPRC suppresses genome instability and hepatocellular carcinomas and promotes survivals in mice by sustaining Yap-P27-mediated cell ploidy and P62-HDAC6-controlled autophagy maturation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: Cancer J Clin. 2015;65:87–108.
Singh S, Singh PP, Roberts LR, Sanchez W. Chemopreventive strategies in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2014;11:45–54.
Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, et al. A protein conjugation system essential for autophagy. Nature. 1998;395:395–8.
Terman A, Gustafsson B, Brunk UT. The lysosomal-mitochondrial axis theory of postmitotic aging and cell death. Chem-Biol Interact. 2006;163:29–37.
Liang J, Shao SH, Xu ZX, Hennessy B, Ding Z, Larrea M, et al. The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat Cell Biol. 2007;9:218–24.
Xie R, Nguyen S, McKeehan WL, Liu L. Acetylated microtubules are required for fusion of autophagosomes with lysosomes. BMC Cell Biol. 2010;11:89.
Marino G, Salvador-Montoliu N, Fueyo A, Knecht E, Mizushima N, Lopez-Otin C. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J Biol Chem. 2007;282:18573–83.
Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Investig. 2003;112:1809–20.
Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800.
Xie R, Wang F, McKeehan WL, Liu L. Autophagy enhanced by microtubule- and mitochondrion-associated MAP1S suppresses genome instability and hepatocarcinogenesis. Cancer Res. 2011;71:7537–46.
Li W, Yue F, Dai Y, Shi B, Xu G, Jiang X, et al. Suppressor of hepatocellular carcinoma RASSF1A activates autophagy initiation and maturation. Cell Death Differ. 2019;26:1379–95.
Liu L, Amy V, Liu G, McKeehan WL. Novel complex integrating mitochondria and the microtubular cytoskeleton with chromosome remodeling and tumor suppressor RASSF1 deduced by in silico homology analysis, interaction cloning in yeast, and colocalization in cultured cells. Vitr Cell Dev Biol Anim. 2002;38:582–94.
Liu L, McKeehan WL. Sequence analysis of LRPPRC and its SEC1 domain interaction partners suggest roles in cytoskeletal organization, vesicular trafficking, nucleocytosolic shuttling and chromosome activity. Genomics. 2002;79:124–36.
Mootha VK, Lepage P, Miller K, Bunkenborg J, Reich M, Hjerrild M, et al. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc Natl Acad Sci USA. 2003;100:605–10.
Ruzzenente B, Metodiev MD, Wredenberg A, Bratic A, Park CB, Camara Y, et al. LRPPRC is necessary for polyadenylation and coordination of translation of mitochondrial mRNAs. EMBO J. 2012;31:443–56.
Xu F, Addis JB, Cameron JM, Robinson BH. LRPPRC mutation suppresses cytochrome oxidase activity by altering mitochondrial RNA transcript stability in a mouse model. Biochem J. 2012;441:275–83.
Liu L, Sanosaka M, Lei S, Bestwick ML, Frey JH Jr, Surovtseva YV, et al. LRP130 protein remodels mitochondria and stimulates fatty acid oxidation. J Biol Chem. 2011;286:41253–64.
Mili S, Pinol-Roma S. LRP130, a pentatricopeptide motif protein with a noncanonical RNA-binding domain, is bound in vivo to mitochondrial and nuclear RNAs. Mol Cell Biol. 2003;23:4972–82.
Siira SJ, Spahr H, Shearwood AJ, Ruzzenente B, Larsson NG, Rackham O, et al. LRPPRC-mediated folding of the mitochondrial transcriptome. Nat Commun. 2017;8:1532.
Nam M, Akie TE, Sanosaka M, Craige SM, Kant S, Keaney JF Jr, et al. Mitochondrial retrograde signaling connects respiratory capacity to thermogenic gene expression. Sci Rep. 2017;7:2013.
Cuillerier A, Honarmand S, Cadete VJJ, Ruiz M, Forest A, Deschenes S, et al. Loss of hepatic LRPPRC alters mitochondrial bioenergetics, regulation of permeability transition and trans-membrane ROS diffusion. Hum Mol Genet. 2017;26:3186–201.
Schweitzer CJ, Matthews JM, Madson CJ, Donnellan MR, Cerny RL, Belshan M. Knockdown of the cellular protein LRPPRC attenuates HIV-1 infection. PLoS ONE. 2012;7:e40537.
Refolo G, Ciccosanti F, Di Rienzo M, Basulto Perdomo A, Romani M, Alonzi T, et al. Negative regulation of mitochondrial antiviral signaling protein-mediated antiviral signaling by the mitochondrial protein LRPPRC during hepatitis C virus infection. Hepatology. 2019;69:34–50.
Liu L, Vo A, Liu G, McKeehan WL. Distinct structural domains within C19ORF5 support association with stabilized microtubules and mitochondrial aggregation and genome destruction. Cancer Res. 2005;65:4191–201.
Liu L, Vo A, McKeehan WL. Specificity of the methylation-suppressed A isoform of candidate tumor suppressor RASSF1 for microtubule hyperstabilization is determined by cell death inducer C19ORF5. Cancer Res. 2005;65:1830–8.
Xie R, Nguyen S, McKeehan K, Wang F, McKeehan WL, Liu L. Microtubule-associated protein 1S (MAP1S) bridges autophagic components with microtubules and mitochondria to affect autophagosomal biogenesis and degradation. J Biol Chem. 2011;286:10367–77.
Xie R. Autophagy enhanced by microtubule- and mitochondrion-associated MAP1S suppresses genome instability and hepatocarcinogenesis. Autophagy. 2011;71:7537–46.
Liu L, McKeehan WL, Wang F, Xie R. MAP1S enhances autophagy to suppress tumorigenesis. Autophagy. 2012;8:278–80.
Liu L, Vo A, Liu G, McKeehan WL. Putative tumor suppressor RASSF1 interactive protein and cell death inducer C19ORF5 is a DNA binding protein. Biochem Biophys Res Commun. 2005;332:670–6.
Jiang X, Li X, Huang H, Jiang F, Lin Z, He H, et al. Elevated levels of mitochondrion-associated autophagy inhibitor LRPPRC are associated with poor prognosis in patients with prostate cancer. Cancer. 2014;120:1228–36.
Zhang S, Chen Q, Liu Q, Li Y, Sun X, Hong L, et al. Hippo signaling suppresses cell ploidy and tumorigenesis through Skp2. Cancer Cell. 2017;31:669–84. e667.
Lee YA, Noon LA, Akat KM, Ybanez MD, Lee TF, Berres ML, et al. Autophagy is a gatekeeper of hepatic differentiation and carcinogenesis by controlling the degradation of Yap. Nat Commun. 2018;9:4962.
Chen Q, Yue F, Li W, Zou J, Xu T, Huang C, et al. Potassium bisperoxo (1,10-phenanthroline) oxovanadate (bpV(phen)) induces apoptosis and pyroptosis and disrupts the P62-HDAC6 interaction to suppress the acetylated microtubule-dependent degradation of autophagosomes. J Biol Chem. 2015;290:26051–8.
Jiang X, Huang Y, Liang X, Jiang F, He Y, Li T, et al. Metastatic prostate cancer-associated P62 inhibits autophagy flux and promotes epithelial to mesenchymal transition by sustaining the level of HDAC6. Prostate. 2018;78:426–34.
Zou J, Yue F, Jiang X, Li W, Yi J, Liu L. Mitochondrion-associated protein LRPPRC suppresses the initiation of basal levels of autophagy via enhancing Bcl-2 stability. Biochem J. 2013;454:447–57.
Zou J, Yue F, Li W, Song K, Jiang X, Yi J, et al. Autophagy inhibitor LRPPRC suppresses mitophagy through interaction with mitophagy initiator Parkin. PLoS ONE. 2014;9:e94903.
Chourasia AH, Boland ML, Macleod KF. Mitophagy and cancer. Cancer Metab. 2015;3:4.
Johansen T, Lamark T. Selective autophagy: ATG8 family proteins, LIR motifs and cargo receptors. J Mol Biol. 2020;432:80–103.
Mishra PK, Raghuram GV, Panwar H, Jain D, Pandey H, Maudar KK. Mitochondrial oxidative stress elicits chromosomal instability after exposure to isocyanates in human kidney epithelial cells. Free Radic Res. 2009;43:718–28.
Li M, Fang X, Baker DJ, Guo L, Gao X, Wei Z, et al. The ATM-p53 pathway suppresses aneuploidy-induced tumorigenesis. Proc Natl Acad Sci USA. 2010;107:14188–93.
Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. 2014;157:1013–22.
Ryter SW, Mizumura K, Choi AM. The impact of autophagy on cell death modalities. Int J Cell Biol. 2014;2014:502676.
Yu J, Nagasu H, Murakami T, Hoang H, Broderick L, Hoffman HM, et al. Inflammasome activation leads to Caspase-1-dependent mitochondrial damage and block of mitophagy. Proc Natl Acad Sci USA. 2014;111:15514–9.
Storchova Z, Pellman D. From polyploidy to aneuploidy, genome instability and cancer. Nat Rev Mol Cell Biol. 2004;5:45–54.
Hergovich A. Mammalian Hippo signalling: a kinase network regulated by protein–protein interactions. Biochem Soc Trans. 2012;40:124–8.
Lai D, Ho KC, Hao Y, Yang X. Taxol resistance in breast cancer cells is mediated by the hippo pathway component TAZ and its downstream transcriptional targets Cyr61 and CTGF. Cancer Res. 2011;71:2728–38.
Chen Q, Xie W, Kuhn DJ, Voorhees PM, Lopez-Girona A, Mendy D, et al. Targeting the p27 E3 ligase SCF(Skp2) results in p27- and Skp2-mediated cell-cycle arrest and activation of autophagy. Blood. 2008;111:4690–9.
Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–8.
Vandin F, Clay P, Upfal E, Raphael BJ. Discovery of mutated subnetworks associated with clinical data in cancer. Pac Symp Biocomput. 2012;2012:55–66.
Xu G, Jiang Y, Xiao Y, Liu XD, Yue F, Li W, et al. Fast clearance of lipid droplets through MAP1S-activated autophagy suppresses clear cell renal cell carcinomas and promotes patient survival. Oncotarget. 2016;7:6255–65.
Yue F, Li W, Zou J, Jiang X, Xu G, Huang H, et al. Spermidine prolongs lifespan and prevents liver fibrosis and hepatocellular carcinoma by activating MAP1S-mediated autophagy. Cancer Res. 2017;77:2938–51.
Li W, Zou J, Yue F, Song K, Chen Q, McKeehan WL, et al. Defects in MAP1S-mediated autophagy cause reduction in mouse lifespans especially when fibronectin is overexpressed. Aging Cell. 2016;15:370–9.
Jiang X, Zhong W, Huang H, He H, Jiang F, Chen Y, et al. Autophagy defects suggested by low levels of autophagy activator MAP1S and high levels of autophagy inhibitor LRPPRC predict poor prognosis of prostate cancer patients. Mol Carcinog. 2015;54:1194–204.
Postic C, Shiota M, Niswender KD, Jetton TL, Chen Y, Moates JM, et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J Biol Chem. 1999;274:305–15.
Yue F, Li W, Zou J, Chen Q, Xu G, Huang H, et al. Blocking the association of HDAC4 with MAP1S accelerates autophagy clearance of mutant Huntingtin. Aging. 2015;7:839–53.
Jimenez-Martinez M, Ostale CM, van der Burg LR, Galan-Martinez J, Hardwick JCH, Lopez-Perez R, et al. DUSP10 is a regulator of YAP1 activity promoting cell proliferation and colorectal cancer progression. Cancers. 2019;11:E1767.
Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Gene Dev. 2007;21:2747–61.
Acknowledgements
We thank Dr Nils-Göran Larsson, Department of Mitochondrial Genetics, Max Planck Institute for Biology of Ageing, Cologne, Germany, for providing LRPPRCloxP/loxP mice to us as a gift. This work was supported by National Natural Science Foundation of China (81772931) and NCI R01CA142862 to Leyuan Liu.
Author information
Authors and Affiliations
Contributions
WL and LL designed the study, interpreted data, and wrote the manuscript. WL, YD, BS, and LL performed the experiments and analyzed data. FY, JZ, GX, XJ, FW, and XZ contributed to experimental design, data interpretation, or research resources. All authors reviewed the manuscript and accepted final version.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Li, W., Dai, Y., Shi, B. et al. LRPPRC sustains Yap-P27-mediated cell ploidy and P62-HDAC6-mediated autophagy maturation and suppresses genome instability and hepatocellular carcinomas. Oncogene 39, 3879–3892 (2020). https://doi.org/10.1038/s41388-020-1257-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-020-1257-9
This article is cited by
-
Pan-cancer evaluation of regulated cell death to predict overall survival and immune checkpoint inhibitor response
npj Precision Oncology (2024)
-
LRPPRC regulates malignant behaviors, protects mitochondrial homeostasis, mitochondrial function in osteosarcoma and derived cancer stem-like cells
BMC Cancer (2023)
-
A traditional gynecological medicine inhibits ovarian cancer progression and eliminates cancer stem cells via the LRPPRC–OXPHOS axis
Journal of Translational Medicine (2023)
-
Comprehensive characterization of m6A methylation and its impact on prognosis, genome instability, and tumor microenvironment in hepatocellular carcinoma
BMC Medical Genomics (2022)
-
LncRNA SNHG17 interacts with LRPPRC to stabilize c-Myc protein and promote G1/S transition and cell proliferation
Cell Death & Disease (2021)