Abstract
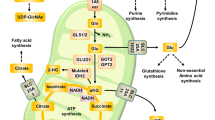
Cancer metabolism research has recently been revived and its focus expanded from glucose and the Warburg’s effects on other nutrients, such as glutamine. The underlying mechanism of oncogenic alterations of glutaminolysis remains unclear. Genetic alterations of EGFR are observed in ~50% of glioblastoma (GBM) patients, and have been found to play important roles in the metabolic abnormalities of GBM. In this study, we found that glutamine metabolism was upregulated after EGFR activation in a GDH1 (glutamate dehydrogenase 1)-dependent manner. Knockdown of GDH1 significantly reduced the cell proliferation, colony formation and tumorigenesis abilities of glioblastoma cells. Furthermore, we showed that GDH1-mediated glutaminolysis was involved in EGF-promoted cell proliferation. EGFR triggered the phosphorylation of ELK1 at Ser 383 through activating MEK/ERK signaling. Phosphorylated ELK1 enriched in the promoter of GDH1 to activate the transcription of GDH1, which then promoted glutamine metabolism. In addition, EGFR activation did not accelerate glutaminolysis in ELK1 knockdown or ELK1 Ser383-mutated cells. Collectively, our findings indicate that EGFR phosphorylates ELK1 to activate GDH1 transcription and glutaminolysis through MEK/ERK pathway, providing new insight into oncogenic alterations of glutamine metabolism.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
12 July 2022
A Correction to this paper has been published: https://doi.org/10.1038/s41388-022-02392-w
References
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74.
DeBerardinis RJ, Cheng T. Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313–24.
Hensley CT, Wasti AT, DeBerardinis RJ. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Investig. 2013;123:3678–84.
Mayers JR, Vander Heiden MG. Famine versus feast: understanding the metabolism of tumors in vivo. Trends Biochemical Sci. 2015;40:130–40.
Altman BJ, Stine ZE, Dang CV. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat Rev Cancer. 2016;16:619–34.
Bhutia YD, Babu E, Ramachandran S, Ganapathy V. Amino Acid transporters in cancer and their relevance to “glutamine addiction”: novel targets for the design of a new class of anticancer drugs. Cancer Res. 2015;75:1782–8.
Jiang L, Shestov AA, Swain P, Yang C, Parker SJ, Wang QA, et al. Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature. 2016;532:255–8.
DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci USA. 2007;104:19345–50.
Fan J, Kamphorst JJ, Mathew R, Chung MK, White E, Shlomi T, et al. Glutamine-driven oxidative phosphorylation is a major ATP source in transformed mammalian cells in both normoxia and hypoxia. Mol Syst Biol. 2013;9:712.
Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–82.
Wang YQ, Wang HL, Xu J, Tan J, Fu LN, Wang JL, et al. Sirtuin5 contributes to colorectal carcinogenesis by enhancing glutaminolysis in a deglutarylation-dependent manner. Nat Commun. 2018;9:545.
Ostrom QT, Gittleman H, Truitt G, Boscia A, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2011-5. Neuro Oncol. 2018;20:iv1–iv86.
Liu F, Hon GC, Villa GR, Turner KM, Ikegami S, Yang H, et al. EGFR mutation promotes glioblastoma through epigenome and transcription factor network remodeling. Mol Cell. 2015;60:307–18.
Halatsch ME, Schmidt U, Behnke-Mursch J, Unterberg A, Wirtz CR. Epidermal growth factor receptor inhibition for the treatment of glioblastoma multiforme and other malignant brain tumours. Cancer Treat Rev. 2006;32:74–89.
Hatanpaa KJ, Burma S, Zhao D, Habib AA. Epidermal growth factor receptor in glioma: signal transduction, neuropathology, imaging, and radioresistance. Neoplasia. 2010;12:675–84.
Felsberg J, Hentschel B, Kaulich K, Gramatzki D, Zacher A, Malzkorn B, et al. Epidermal growth factor receptor variant III (EGFRvIII) positivity in EGFR-amplified glioblastomas: prognostic role and comparison between primary and recurrent tumors. Clin Cancer Res. 2017;23:6846–55.
Babic I, Anderson ES, Tanaka K, Guo D, Masui K, Li B, et al. EGFR mutation-induced alternative splicing of Max contributes to growth of glycolytic tumors in brain cancer. Cell Metab. 2013;17:1000–8.
Mai WX, Gosa L, Daniels VW, Ta L, Tsang JE, Higgins B, et al. Cytoplasmic p53 couples oncogene-driven glucose metabolism to apoptosis and is a therapeutic target in glioblastoma. Nat Med. 2017;23:1342–51.
Ji H, Lee JH, Wang Y, Pang Y, Zhang T, Xia Y, et al. EGFR phosphorylates FAM129B to promote Ras activation. Proc Natl Acad Sci USA. 2016;113:644–9.
Lee JH, Liu R, Li J, Wang Y, Tan L, Li XJ, et al. EGFR-phosphorylated platelet isoform of phosphofructokinase 1 promotes PI3K activation. Mol Cell. 2018;70:197–210.e197.
Jin L, Li D, Alesi GN, Fan J, Kang HB, Lu Z, et al. Glutamate dehydrogenase 1 signals through antioxidant glutathione peroxidase 1 to regulate redox homeostasis and tumor growth. Cancer Cell. 2015;27:257–70.
Yordy JS, Muise-Helmericks RC. Signal transduction and the Ets family of transcription factors. Oncogene. 2000;19:6503–13.
Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74.
Jameson KL, Mazur PK, Zehnder AM, Zhang J, Zarnegar B, Sage J, et al. IQGAP1 scaffold-kinase interaction blockade selectively targets RAS-MAP kinase-driven tumors. Nat Med. 2013;19:626–30.
Agnihotri S, Zadeh G. Metabolic reprogramming in glioblastoma: the influence of cancer metabolism on epigenetics and unanswered questions. Neuro Oncol. 2016;18:160–72.
Tateishi K, Iafrate AJ, Ho Q, Curry WT, Batchelor TT, Flaherty KT, et al. Myc-driven glycolysis is a therapeutic target in glioblastoma. Clin Cancer Res. 2016;22:4452–65.
Michalak KP, Mackowska-Kedziora A, Sobolewski B, Wozniak P. Key roles of glutamine pathways in reprogramming the cancer metabolism. Oxid Med Cell Longev. 2015;2015:964321.
Tanaka K, Sasayama T, Irino Y, Takata K, Nagashima H, Satoh N, et al. Compensatory glutamine metabolism promotes glioblastoma resistance to mTOR inhibitor treatment. J Clin Investig. 2015;125:1591–602.
Stehle P, Ellger B, Kojic D, Feuersenger A, Schneid C, Stover J, et al. Glutamine dipeptide-supplemented parenteral nutrition improves the clinical outcomes of critically ill patients: a systematic evaluation of randomised controlled trials. Clin Nutr ESPEN. 2017;17:75–85.
Kovacevic Z. The pathway of glutamine and glutamate oxidation in isolated mitochondria from mammalian cells. Biochemical J. 1971;125:757–63.
Craze ML, El-Ansari R, Aleskandarany MA, Cheng KW, Alfarsi L, Masisi B, et al. Glutamate dehydrogenase (GLUD1) expression in breast cancer. Breast Cancer Res Treat. 2019;174:79–91.
Nwosu ZC, Battello N, Rothley M, Pioronska W, Sitek B, Ebert MP, et al. Liver cancer cell lines distinctly mimic the metabolic gene expression pattern of the corresponding human tumours. J Exp Clin Cancer Res. 2018;37:211.
Jin L, Chun J, Pan C, Kumar A, Zhang G, Ha Y, et al. The PLAG1-GDH1 axis promotes anoikis resistance and tumor metastasis through CamKK2-AMPK signaling in LKB1-deficient lung cancer. Mol Cell. 2018;69:87–99.e87.
Shao N, Chai Y, Cui JQ, Wang N, Aysola K, Reddy ES, et al. Induction of apoptosis by Elk-1 and deltaElk-1 proteins. Oncogene. 1998;17:527–32.
Chai Y, Chipitsyna G, Cui J, Liao B, Liu S, Aysola K, et al. c-Fos oncogene regulator Elk-1 interacts with BRCA1 splice variants BRCA1a/1b and enhances BRCA1a/1b-mediated growth suppression in breast cancer cells. Oncogene. 2001;20:1357–67.
Zhang X, Zhang B, Gao J, Wang X, Liu Z. Regulation of the microRNA 200b (miRNA-200b) by transcriptional regulators PEA3 and ELK-1 protein affects expression of Pin1 protein to control anoikis. J Biol Chem. 2013;288:32742–52.
Dittmer J, Nordheim A. Ets transcription factors and human disease. Biochim Biophys Acta. 1998;1377:F1–11.
Ma J, Zeng S, Zhang Y, Deng G, Qu Y, Guo C, et al. BMP4 promotes oxaliplatin resistance by an induction of epithelial-mesenchymal transition via MEK1/ERK/ELK1 signaling in hepatocellular carcinoma. Cancer Lett. 2017;411:117–29.
Ohguchi H, Harada T, Sagawa M, Kikuchi S, Tai YT, Richardson PG, et al. KDM6B modulates MAPK pathway mediating multiple myeloma cell growth and survival. Leukemia. 2017;31:2661–9.
Hipskind RA, Rao VN, Mueller CG, Reddy ES, Nordheim A. Ets-related protein Elk-1 is homologous to the c-fos regulatory factor p62TCF. Nature. 1991;354:531–4.
Yeh HY, Cheng SW, Lin YC, Yeh CY, Lin SF, Soo VW. Identifying significant genetic regulatory networks in the prostate cancer from microarray data based on transcription factor analysis and conditional independency. BMC Med Genom. 2009;2:70.
Maicas M, Vazquez I, Vicente C, Garcia-Sanchez MA, Marcotegui N, Urquiza L, et al. Functional characterization of the promoter region of the human EVI1 gene in acute myeloid leukemia: RUNX1 and ELK1 directly regulate its transcription. Oncogene. 2013;32:2069–78.
Boros J, Donaldson IJ, O’Donnell A, Odrowaz ZA, Zeef L, Lupien M, et al. Elucidation of the ELK1 target gene network reveals a role in the coordinate regulation of core components of the gene regulation machinery. Genome Res. 2009;19:1963–73.
Yang R, Yi L, Dong Z, Ouyang Q, Zhou J, Pang Y, et al. Tigecycline inhibits glioma growth by regulating miRNA-199b-5p-HES1-AKT pathway. Mol Cancer Therapeutics. 2016;15:421–9.
Acknowledgements
This research was supported by the National Natural Science Foundation of China (Nos. 81672502 and 81872071), the grant to the Key Laboratory of Higher Education Institutes of Shandong Province, the National Natural Science Foundation Cultivation Project of Jining Medical University (600788002) and Faculty Startup Fund for RY from Jining Medical University (600788001).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Yang, R., Li, X., Wu, Y. et al. EGFR activates GDH1 transcription to promote glutamine metabolism through MEK/ERK/ELK1 pathway in glioblastoma. Oncogene 39, 2975–2986 (2020). https://doi.org/10.1038/s41388-020-1199-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-020-1199-2
This article is cited by
-
A resource database for protein kinase substrate sequence-preference motifs based on large-scale mass spectrometry data
Cell Communication and Signaling (2024)
-
Circular RNA Dipeptidyl Peptidase 4 (circDPP4) Stimulates the Expression of Glutamate Dehydrogenase 1 to Contribute to the Malignant Phenotypes of Prostate Cancer by Sponging miR-497-5p
Molecular Biotechnology (2024)
-
Toward the next generation EGFR inhibitors: an overview of osimertinib resistance mediated by EGFR mutations in non-small cell lung cancer
Cell Communication and Signaling (2023)
-
TFEB inhibition induces melanoma shut-down by blocking the cell cycle and rewiring metabolism
Cell Death & Disease (2023)
-
Extracellular vesicles derived from glioblastoma promote proliferation and migration of neural progenitor cells via PI3K-Akt pathway
Cell Communication and Signaling (2022)