Abstract
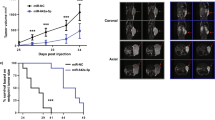
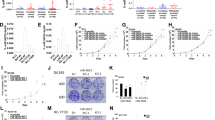
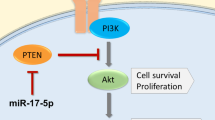
The chromosome 8q24.21 locus, which contains the proto-oncogene c-MYC, long non-coding RNA PVT1, and microRNAs (miRs), is the most commonly amplified region in human prostate cancer. A long-range interaction of genetic variants with c-MYC or long non-coding PVT1 at this locus contributes to the genetic risk of prostate cancer. At this locus is a cluster of genes for six miRs (miR-1204, -1205, -1206, -1207-3p, -1207-5p, and -1208), but their functional role remains elusive. Here the copy numbers and expression levels of miRs-1204–1208 were investigated using quantitative PCR for prostate cancer cell lines and primary tumors. The data revealed that copy numbers and expression of miR-1205 were increased in both castration-resistant prostate cancer cell lines and in primary tumors. In castration-resistant prostate cancer specimens, the copy number at the miR-1205 locus correlated with the expression of miR-1205. Furthermore, functional analysis with an miR-1205 mimic, an miR-1205 inhibitor, and CRISPR/Cas9 knockout revealed that, in human prostate cancer cells, miR-1205 promoted cell proliferation and cell cycle progression and inhibited hydrogen peroxide-induced apoptosis. In these cells, miR-1205 downregulated the expression of the Egl-9 family hypoxia inducible factor 3(EGLN3) gene and targeted a site in its 3′-untranslated region to downregulate its transcriptional activity. Thus, by targeting EGLN3, miR-1205 has an oncogenic role and may contribute to the genetic risk of castration-resistant prostate cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30.
Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, Helgason A, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–7.
Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–9.
Eeles RA, Kote-Jarai Z, Giles GG, Olama AA, Guy M, Jugurnauth SK, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–21.
Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–5.
Ahmadiyeh N, Pomerantz MM, Grisanzio C, Herman P, Jia L, Almendro V, et al. 8q24 prostate, breast, and colon cancer risk loci show tissue-specific long-range interaction with MYC. Proc Natl Acad Sci USA. 2010;107:9742–6.
Kim T, Cui R, Jeon YJ, Lee JH, Lee JH, Sim H, et al. Long-range interaction and correlation between MYC enhancer and oncogenic long noncoding RNA CARLo-5. Proc Natl Acad Sci USA. 2014;111:4173–8.
Meyer KB, Maia AT, O’Reilly M, Ghoussaini M, Prathalingam R, Porter-Gill P, et al. A functional variant at a prostate cancer predisposition locus at 8q24 is associated with PVT1 expression. PLoS Genet. 2011;7:e1002165.
Cher ML, MacGrogan D, Bookstein R, Brown JA, Jenkins RB, Jensen RH. Comparative genomic hybridization, allelic imbalance, and fluorescence in situ hybridization on chromosome 8 in prostate cancer. Genes Chromosomes Cancer. 1994;11:153–62.
Visakorpi T, Kallioniemi AH, Syvanen AC, Hyytinen ER, Karhu R, Tammela T, et al. Genetic changes in primary and recurrent prostate cancer by comparative genomic hybridization. Cancer Res. 1995;55:342–7.
Van Den Berg C, Guan XY, Von Hoff D, Jenkins R, Bittner, Griffin C, et al. DNA sequence amplification in human prostate cancer identified by chromosome microdissection: potential prognostic implications. Clin Cancer Res. 1995;1:11–18.
Tseng YY, Moriarity BS, Gong W, Akiyama R, Tiwari A, Kawakami H, et al. PVT1 dependence in cancer with MYC copy-number increase. Nature. 2014;512:82–86.
Huppi K, Pitt JJ, Wahlberg BM, Caplen NJ. The 8q24 gene desert: an oasis of non-coding transcriptional activity. Front Genet. 2012;3:69.
Beck-Engeser GB, Lum AM, Huppi K, Caplen NJ, Wang BB, Wabl M. Pvt1-encoded microRNAs in oncogenesis. Retrovirology. 2008;5:4.
Huppi K, Volfovsky N, Runfola T, Jones TL, Mackiewicz M, Martin SE, et al. The identification of microRNAs in a genomically unstable region of human chromosome 8q24. Mol Cancer Res. 2008;6:212–21.
Sato K, Qian J, Slezak JM, Lieber MM, Bostwick DG, Bergstralh EJ, et al. Clinical significance of alterations of chromosome 8 in high-grade, advanced, nonmetastatic prostate carcinoma. J Natl Cancer Inst. 1999;91:1574–80.
Lapointe J, Li C, Giacomini CP, Salari K, Huang S, Wang P, et al. Genomic profiling reveals alternative genetic pathways of prostate tumorigenesis. Cancer Res. 2007;67:8504–10.
Bawa P, Zackaria S, Verma M, Gupta S, Srivatsan R, Chaudhary B, et al. Integrative analysis of normal long intergenic non-coding RNAs in prostate cancer. PLoS ONE. 2015;10:e0122143.
Cho SW, Xu J, Sun R, Mumbach MR, Carter AC, Chen YG, et al. Promoter of lncRNA gene PVT1 is a tumor-suppressor DNA boundary element. Cell. 2018;173:1398–412.
The PVT1 promoter suppresses MYC transcription to reduce cell growth. Cancer Discov. 2018;8:OF15.
Tang J, Li Y, Sang Y, Yu B, Lv D, Zhang W. et al. LncRNA PVT1 regulates triple-negative breast cancer through KLF5/beta-catenin signaling. Oncogene. 2018;37:4723–34.
Zhao J, Du P, Cui P, Qin Y, Hu C, Wu J. et al. LncRNA PVT1 promotes angiogenesis via activating the STAT3/VEGFA axis in gastric cancer. Oncogene. 2018;37:4094–109.
Barsotti AM, Beckerman R, Laptenko O, Huppi K, Caplen NJ, Prives C. p53-Dependent induction of PVT1 and miR-1204. J Biol Chem. 2012;287:2509–19.
Peng X, Cao P, Li J, He D, Han S, Zhou J, et al. MiR-1204 sensitizes nasopharyngeal carcinoma cells to paclitaxel both in vitro and in vivo. Cancer Biol Ther. 2015;16:261–7.
Liu X, Bi L, Wang Q, Wen M, Li C, Ren Y. et al. miR-1204 targets VDR to promotes epithelial-mesenchymal transition and metastasis in breast cancer. Oncogene. 2018;37:3426–39.
Riquelme E, Suraokar MB, Rodriguez J, Mino B, Lin HY, Rice DC, et al. Frequent coamplification and cooperation between C-MYC and PVT1 oncogenes promote malignant pleural mesothelioma. J Thorac Oncol. 2014;9:998–1007.
Wu G, Liu A, Zhu J, Lei F, Wu S, Zhang X, et al. MiR-1207 overexpression promotes cancer stem cell-like traits in ovarian cancer by activating the Wnt/beta-catenin signaling pathway. Oncotarget. 2015;6:28882–94.
Yan C, Chen Y, Kong W, Fu L, Liu Y, Yao Q, et al. PVT1-derived miR-1207-5p promotes breast cancer cell growth by targeting STAT6. Cancer Sci. 2017;108:868–76.
Das DK, Naidoo M, Ilboudo A, Park JY, Ali T, Krampis K, et al. miR-1207-3p regulates the androgen receptor in prostate cancer via FNDC1/fibronectin. Exp Cell Res. 2016;348:190–200.
Sato H, Minei S, Hachiya T, Yoshida T, Takimoto Y. Fluorescence in situ hybridization analysis of c-myc amplification in stage TNM prostate cancer in Japanese patients. Int J Urol. 2006;13:761–6.
Sotelo J, Esposito D, Duhagon MA, Banfield K, Mehalko J, Liao H, et al. Long-range enhancers on 8q24 regulate c-Myc. Proc Natl Acad Sci USA. 2010;107:3001–5.
Wasserman NF, Aneas I, Nobrega MA. An 8q24 gene desert variant associated with prostate cancer risk confers differential in vivo activity to a MYC enhancer. Genome Res. 2010;20:1191–7.
Weijerman PC, Konig JJ, Wong ST, Niesters HG, Peehl DM. Lipofection-mediated immortalization of human prostatic epithelial cells of normal and malignant origin using human papillomavirus type 18 DNA. Cancer Res. 1994;54:5579–83.
Shi Y, Yang F, Wei S, Xu G. Identification of key genes affecting results of hyperthermia in osteosarcoma based on integrative ChIP-Seq/TargetScan analysis. Med Sci Monit. 2017;23:2042–8.
min H, Yoon S. Got target? Computational methods for microRNA target prediction and their extension. Exp Mol Med. 2010;42:233–44.
Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43:D146–152.
Luo W, Lin B, Wang Y, Zhong J, O’Meally R, Cole RN, et al. PHD3-mediated prolyl hydroxylation of nonmuscle actin impairs polymerization and cell motility. Mol Biol Cell. 2014;25:2788–96.
Shi H, Zheng B, Wu Y, Tang Y, Wang L, Gao Y, et al. Ubiquitin ligase Siah1 promotes the migration and invasion of human glioma cells by regulating HIF-1alpha signaling under hypoxia. Oncol Rep. 2015;33:1185–90.
Garvalov BK, Foss F, Henze AT, Bethani I, Graf-Hochst S, Singh D, et al. PHD3 regulates EGFR internalization and signalling in tumours. Nat Commun. 2014;5:5577.
Henze AT, Garvalov BK, Seidel S, Cuesta AM, Ritter M, Filatova A, et al. Loss of PHD3 allows tumours to overcome hypoxic growth inhibition and sustain proliferation through EGFR. Nat Commun. 2014;5:5582.
Place TL, Fitzgerald MP, Venkataraman S, Vorrink SU, Case AJ, Teoh ML, et al. Aberrant promoter CpG methylation is a mechanism for impaired PHD3 expression in a diverse set of malignant cells. PLoS ONE. 2011;6:e14617.
Lee S, Nakamura E, Yang H, Wei W, Linggi MS, Sajan MP, et al. Neuronal apoptosis linked to EglN3 prolyl hydroxylase and familial pheochromocytoma genes: developmental culling and cancer. Cancer Cell. 2005;8:155–67.
Schlisio S. Neuronal apoptosis by prolyl hydroxylation: implication in nervous system tumours and the Warburg conundrum. J Cell Mol Med. 2009;13:4104–12.
Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discov. 2009;8:579–91.
Vergara R, Parada F, Rubio S, Perez FJ. Hypoxia induces H2O2 production and activates antioxidant defence system in grapevine buds through mediation of H2O2 and ethylene. J Exp Bot. 2012;63:4123–31.
Kaelin WG Jr., Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402.
Fu J, Taubman MB. EGLN3 inhibition of NF-kappaB is mediated by prolyl hydroxylase-independent inhibition of IkappaB kinase gamma ubiquitination. Mol Cell Biol. 2013;33:3050–61.
Lipscomb EA, Sarmiere PD, Crowder RJ, Freeman RS. Expression of the SM-20 gene promotes death in nerve growth factor-dependent sympathetic neurons. J Neurochem. 1999;73:429–32.
Madden SL, Galella EA, Riley D, Bertelsen AH, Beaudry GA. Induction of cell growth regulatory genes by p53. Cancer Res. 1996;56:5384–90.
Gerber SA, Yatsula B, Maier CL, Sadler TJ, Whittaker LW, Pober JS. Interferon-gamma induces prolyl hydroxylase (PHD)3 through a STAT1-dependent mechanism in human endothelial cells. Arterioscler Thromb Vasc Biol. 2009;29:1363–9.
Hogel H, Rantanen K, Jokilehto T, Grenman R, Jaakkola PM. Prolyl hydroxylase PHD3 enhances the hypoxic survival and G1 to S transition of carcinoma cells. PLoS ONE. 2011;6:e27112.
Hogel H, Miikkulainen P, Bino L, Jaakkola PM. Hypoxia inducible prolyl hydroxylase PHD3 maintains carcinoma cell growth by decreasing the stability of p27. Mol Cancer. 2015;14:143.
Rodriguez J, Herrero A, Li S, Rauch N, Quintanilla A, Wynne K, et al. PHD3 regulates p53 protein stability by hydroxylating proline 359. Cell Rep. 2018;24:1316–29.
Zhang G, Zhang W, Li B, Stringer-Reasor E, Chu C, Sun L, et al. MicroRNA-200c and microRNA- 141 are regulated by a FOXP3-KAT2B axis and associated with tumor metastasis in breast cancer. Breast Cancer Res. 2017;19:73.
Liu R, Yi B, Wei S, Yang WH, Hart KM, Chauhan P, et al. FOXP3-miR-146-NF-kappaB axis and therapy for precancerous lesions in prostate. Cancer Res. 2015;75:1714–24.
Wang L, Liu R, Li W, Chen C, Katoh H, Chen GY, et al. Somatic single hits inactivate the X-linked tumor suppressor FOXP3 in the prostate. Cancer Cell. 2009;16:336–46.
Zhang W, Yi B, Wang C, Chen D, Bae S, Wei S, et al. Silencing of CD24 enhances the PRIMA-1-induced restoration of mutant p53 in prostate cancer cells. Clin Cancer Res. 2016;22:2545–54.
Ma L, Chung WK. Quantitative analysis of copy number variants based on real-time LightCycler PCR. Curr Protoc Hum Genet. 2014;80:Unit 7.21.
Luo Y, Li B, Zhang G, He Y, Bae JH, Hu F, et al. Integrated oncogenomic profiling of copy numbers and gene expression in lung adenocarcinomas without EGFR mutations or ALK fusion. J Cancer. 2018;9:1096–105.
Gao S, Wang Y, Wang M, Li Z, Zhao Z, Wang RX, et al. MicroRNA-155, induced by FOXP3 through transcriptional repression of BRCA1, is associated with tumor initiation in human breast cancer. Oncotarget. 2017;8:41451–64.
Wang L, Liu R, Ye P, Wong C, Chen GY, Zhou P, et al. Intracellular CD24 disrupts the ARF-NPM interaction and enables mutational and viral oncogene-mediated p53 inactivation. Nat Commun. 2015;6:5909.
Liu R, Liu C, Chen D, Yang WH, Liu X, Liu CG, et al. FOXP3 controls an miR-146/NF-kappaB negative feedback loop that inhibits apoptosis in breast cancer cells. Cancer Res. 2015;75:1703–13.
Liu R, Wang L, Chen G, Katoh H, Chen C, Liu Y, et al. FOXP3 up-regulates p21 expression by site-specific inhibition of histone deacetylase 2/histone deacetylase 4 association to the locus. Cancer Res. 2009;69:2252–9.
Bae S, Park J, Kim JS. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–5.
Acknowledgements
We thank Dr. Donald L Hill for editorial assistance in preparing this manuscript. This work was supported by grants from the National Institutes of Health/National Cancer Institute (CA118948, CA179282, and CA013148 to LW and RL), the Department of Defense (BC160808, PC130594, and PC140308 to LW and RL), the Mike Slive Foundation for Prostate Cancer Research (to LW), and the Natural Science Foundation of China (Nos. 31571126, 31571342, and 81772757 to XL, BL, and RC). Results are based, in part, on data generated by the TCGA Research Network: http://cancergenome.nih.gov/.
Author information
Authors and Affiliations
Contributions
All authors read and approved the final manuscript. RL and RC designed the studies; YW, WL, XL, XML, RL, and LW carried out the experiments; RL, BL, YW, and LW wrote the manuscript; and LW, YW, BL, XL, XML, RC, and RL performed data analysis.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Wang, Y., Li, X., Liu, W. et al. MicroRNA-1205, encoded on chromosome 8q24, targets EGLN3 to induce cell growth and contributes to risk of castration-resistant prostate cancer. Oncogene 38, 4820–4834 (2019). https://doi.org/10.1038/s41388-019-0760-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-019-0760-3
This article is cited by
-
Acquired copy number variation in prostate tumours: a review of common somatic copy number alterations, how they are formed and their clinical utility
British Journal of Cancer (2024)
-
Circular RNA circPTPRF promotes the progression of GBM via sponging miR-1208 to up-regulate YY1
Cancer Cell International (2022)
-
TUBB4A interacts with MYH9 to protect the nucleus during cell migration and promotes prostate cancer via GSK3β/β-catenin signalling
Nature Communications (2022)
-
The microRNA-3622 family at the 8p21 locus exerts oncogenic effects by regulating the p53-downstream gene network in prostate cancer progression
Oncogene (2022)
-
CRISPR interference and activation of the microRNA-3662-HBP1 axis control progression of triple-negative breast cancer
Oncogene (2022)