Abstract
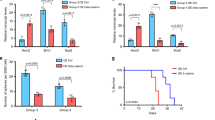
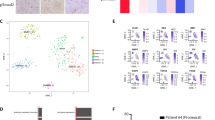
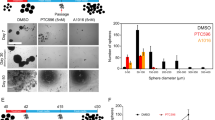
Medulloblastoma (MB) is the most frequent malignant pediatric brain tumor, representing 20% of newly diagnosed childhood central nervous system malignancies. Although advances in multimodal therapy yielded a 5-year survivorship of 80%, MB still accounts for the leading cause of childhood cancer mortality. In this work, we describe the epigenetic regulator BMI1 as a novel therapeutic target for the treatment of recurrent human Group 3 MB, a childhood brain tumor for which there is virtually no treatment option beyond palliation. Current clinical trials for recurrent MB patients based on genomic profiles of primary, treatment-naive tumors will provide limited clinical benefit since recurrent metastatic MBs are highly genetically divergent from their primary tumor. Using a small molecule inhibitor against BMI1, PTC-028, we were able to demonstrate complete ablation of self-renewal of MB stem cells in vitro. When administered to mice xenografted with patient tumors, we observed significant reduction in tumor burden in both local and metastatic compartments and subsequent increased survival, without neurotoxicity. Strikingly, serial in vivo re-transplantation assays demonstrated a marked reduction in tumor initiation ability of recurrent MB cells upon re-transplantation of PTC-028-treated cells into secondary recipient mouse brains. As Group 3 MB is often metastatic and uniformly fatal at recurrence, with no current or planned trials of targeted therapy, an efficacious targeted agent would be rapidly transitioned to clinical trials.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Ellison DW. Childhood medulloblastoma: novel approaches to the classification of a heterogeneous disease. Acta Neuropathol. 2010;120:305–16.
Huse JT, Holland EC. Targeting brain cancer: advances in the molecular pathology of malignant glioma and medulloblastoma. Nat Rev Cancer. 2010;10:319–31.
Taylor MD, Northcott PA, Korshunov A, Remke M, Cho YJ, Clifford SC, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123:465–72.
Cho JH, Wang K, Galas DJ. An integrative approach to inferring biologically meaningful gene modules. BMC Syst Biol. 2011;5:117.
Kool M, Koster J, Bunt J, Hasselt NE, Lakeman A, van Sluis P, et al. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS ONE. 2008;3:e3088.
Northcott PA, Korshunov A, Witt H, Hielscher T, Eberhart CG, Mack S, et al. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol. 2011;29:1408–14.
Pomeroy SL, Tamayo P, Gaasenbeek M, Sturla LM, Angelo M, McLaughlin ME, et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature. 2002;415:436–42.
Thompson MC, Fuller C, Hogg TL, Dalton J, Finkelstein D, Lau CC, et al. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J Clin Oncol. 2006;24:1924–31.
Cavalli FMG, Remke M, Rampasek L, Peacock J, Shih DJH, Luu B, et al. Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell. 2017;31:737–54 e6.
Schwalbe EC, Lindsey JC, Nakjang S, Crosier S, Smith AJ, Hicks D, et al. Novel molecular subgroups for clinical classification and outcome prediction in childhood medulloblastoma: a cohort study. Lancet Oncol. 2017;18:958–71.
Ramaswamy V, Remke M, Bouffet E, Faria CC, Perreault S, Cho YJ, et al. Recurrence patterns across medulloblastoma subgroups: an integrated clinical and molecular analysis. Lancet Oncol. 2013;14:1200–7.
Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–11.
Sauvageau M, Sauvageau G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2010;7:299–313.
Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–56.
Alkema MJ, Wiegant J, Raap AK, Berns A, van Lohuizen M. Characterization and chromosomal localization of the human proto-oncogene BMI-1. Hum Mol Genet. 1993;2:1597–603.
Bruggeman SW, Hulsman D, Tanger E, Buckle T, Blom M, Zevenhoven J, et al. Bmi1 controls tumor development in an Ink4a/Arf-independent manner in a mouse model for glioma. Cancer Cell. 2007;12:328–41.
Gargiulo G, Cesaroni M, Serresi M, de Vries N, Hulsman D, Bruggeman SW, et al. In vivo RNAi screen for BMI1 targets identifies TGF-beta/BMP-ER stress pathways as key regulators of neural- and malignant glioma-stem cell homeostasis. Cancer Cell. 2013;23:660–76.
Leung C, Lingbeek M, Shakhova O, Liu J, Tanger E, Saremaslani P, et al. Bmi1 is essential for cerebellar development and is overexpressed in human medulloblastomas. Nature. 2004;428:337–41.
Wang X, Venugopal C, Manoranjan B, McFarlane N, O'Farrell E, Nolte S, et al. Sonic hedgehog regulates Bmi1 in human medulloblastoma brain tumor-initiating cells. Oncogene. 2012;31:187–99.
Kreso A, van Galen P, Pedley NM, Lima-Fernandes E, Frelin C, Davis T, et al. Self-renewal as a therapeutic target in human colorectal cancer. Nat Med. 2014;20:29–36.
Yong KJ, Basseres DS, Welner RS, Zhang WC, Yang H, Yan B, et al. Targeted BMI1 inhibition impairs tumor growth in lung adenocarcinomas with low CEBPalpha expression. Sci Transl Med. 2016;8:350ra104.
Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest. 2005;115:1503–21.
Wang X, Dubuc AM, Ramaswamy V, Mack S, Gendoo DM, Remke M, et al. Medulloblastoma subgroups remain stable across primary and metastatic compartments. Acta Neuropathol. 2015;129:449–57.
Facchino S, Abdouh M, Chatoo W, Bernier G. BMI1 confers radioresistance to normal and cancerous neural stem cells through recruitment of the DNA damage response machinery. J Neurosci. 2010;30:10096–111.
Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–8.
Wu X, Northcott PA, Dubuc A, Dupuy AJ, Shih DJ, Witt H, et al. Clonal selection drives genetic divergence of metastatic medulloblastoma. Nature. 2012;482:529–33.
Ismail IH, Andrin C, McDonald D, Hendzel MJ. BMI1-mediated histone ubiquitylation promotes DNA double-strand break repair. J Cell Biol. 2010;191:45–60.
Pei Y, Moore CE, Wang J, Tewari AK, Eroshkin A, Cho YJ, et al. An animal model of MYC-driven medulloblastoma. Cancer Cell. 2012;21:155–67.
Bandopadhayay P, Bergthold G, Nguyen B, Schubert S, Gholamin S, Tang Y, et al. BET bromodomain inhibition of MYC-amplified medulloblastoma. Clin Cancer Res. 2014;20:912–25.
Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–6.
Venugopal C, Hallett R, Vora P, Manoranjan B, Mahendram S, Qazi MA, et al. Pyrvinium targets CD133 in human glioblastoma brain tumor-initiating cells. Clin Cancer Res. 2015;21:5324–37.
He XM, Wikstrand CJ, Friedman HS, Bigner SH, Pleasure S, Trojanowski JQ, et al. Differentiation characteristics of newly established medulloblastoma cell lines (D384 Med, D425 Med, and D458 Med) and their transplantable xenografts. Lab Invest. 1991;64:833–43.
Venugopal C, Wang XS, Manoranjan B, McFarlane N, Nolte S, Li M, et al. GBM secretome induces transient transformation of human neural precursor cells. J Neurooncol. 2012;109:457–66.
Tropepe V, Sibilia M, Ciruna BG, Rossant J, Wagner EF, van der Kooy D. Distinct neural stem cells proliferate in response to EGF and FGF in the developing mouse telencephalon. Dev Biol. 1999;208:166–88.
Hallett RM, Dvorkin-Gheva A, Bane A, Hassell JA. A gene signature for predicting outcome in patients with basal-like breast cancer. Sci Rep. 2012;2:227.
Hallett RM, Pond G, Hassell JA. A target based approach identifies genomic predictors of breast cancer patient response to chemotherapy. BMC Med Genom. 2012;5:16.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50.
Cho YJ, Tsherniak A, Tamayo P, Santagata S, Ligon A, Greulich H, et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J Clin Oncol. 2011;29:1424–30.
Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64.
Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401.
Acknowledgements
SKS is supported by Canada Research Chair award, and operating grants from Canadian Institutes of Health Research (CIHR), Stem Cell Network, the Ontario Institute for Cancer Research Cancer Stem Cell Program, the Canadian Cancer Society Research Institute, the Cancer Research Society, the Brain Tumor Foundation of Canada, and generous donations from the Box Run Foundation, Team Kelsey, and patients and their families.
Author contributions
DB: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript. CV and AAA: conception and design, collection and/or assembly of data, manuscript writing, final approval of manuscript. NG and BM: collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript. RH, XW, S Mahendram, PV, TV, M Subapanditha, M Singh, MMK-S, MQ, NM and AM: data analysis and interpretation, final approval of manuscript. OAA and BY: provision of study material or patients, final approval of manuscript. VR, HF and S Morrissy: collection and/or assembly of data, data analysis and interpretation. LC, NS, RB, WD, JS, MW, Y-CM and C-SL: provision of study material or patients, final approval of manuscript. JMK, KHD: data analysis and interpretation. BD, Y-JC, S Mitra, DK and MDT: conception and design, data analysis and interpretation, final approval of manuscript. TWD: provision of study material or patients, final approval of manuscript. SKS: conception and design, data analysis and interpretation, manuscript writing, final approval of manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Bakhshinyan, D., Venugopal, C., Adile, A.A. et al. BMI1 is a therapeutic target in recurrent medulloblastoma. Oncogene 38, 1702–1716 (2019). https://doi.org/10.1038/s41388-018-0549-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-018-0549-9
This article is cited by
-
A prognostic model for Schistosoma japonicum infection-associated liver hepatocellular carcinoma: strengthening the connection through initial biological experiments
Infectious Agents and Cancer (2024)
-
De novo necroptosis creates an inflammatory environment mediating tumor susceptibility to immune checkpoint inhibitors
Communications Biology (2020)
-
Wnt activation as a therapeutic strategy in medulloblastoma
Nature Communications (2020)