Abstract
Microbial interaction with the host through sensing receptors, including SIGNR1, sustains intestinal homeostasis against pathogenic inflammation. The newly discovered commensal Propionibacterium strain, P. UF1, regulates the intestinal immunity against pathogen challenge. However, the molecular events driving intestinal phagocytic cell response, including colonic dendritic cells (DCs), by this bacterium are still elusive. Here, we demonstrate that the glycosylation of bacterial large surface layer protein A (LspA) by protein O-mannosyltransferase 1 (Pmt1) regulates the interaction with SIGNR1, resulting in the control of DC transcriptomic and metabolomic machineries. Programmed DCs promote protective T cell response to intestinal Listeria infection and resist chemically induced colitis in mice. Thus, our findings may highlight a novel molecular mechanism by which commensal surface glycosylation interacting with SIGNR1 directs the intestinal homeostasis to potentially protect the host against proinflammatory signals inducing colonic tissue damage.
Similar content being viewed by others
Introduction
Commensal bacteria, via their surface layer (S-layer) gene products, and the gastrointestinal phagocytic cells expressing sensing receptors (e.g., SIGNR1) synergistically interact to fine-tune the T cell signaling that is critical for protecting the host against pathogenic inflammation exerted by intestinal infections.1,2 In this process, bacterial S-layer macromolecules along with induced metabolites transduce critical signals via cognate receptors into these cells that profoundly control the host homeostasis to protect against tissue damage.3,4 Although the bacterial S-layer proteins display a similar architecture composed of a peptidoglycan layer decorated with proteins and polysaccharides, various modifications, particularly glycosylation, exhibit strain-specific properties that differentially modify the host immune physiology.5 Disruption of mutualistic interactions of the commensal’s S-layer with the host triggers deleterious signals that may manifest in pathogenic inflammation potentially impairing the intestinal barrier function.6 Thus, understanding how host intestinal immunity is regulated through the recognition of these well-structured bacterial gene products by their cognate receptors7 to coordinate protective immune responses is currently of particular therapeutic significance and requires further mechanistic investigations.8
Propionibacterium strain, P. UF1, is a newly discovered commensal bacterium isolated from the gut microbiota of premature infants fed human breast milk.9 This bacterium increases the frequency of colonic Th17 and Treg cells9 involved in mucosal barrier repair and regulation of the intestinal inflammation.10 Induced bacteria-specific Th17 cell differentiation requires the bacterial dihydrolipoamide acetyltransferase (DlaT), an enzymatic component of the pyruvate dehydrogenase complex.9 Chromosomal deletion of dlaT gene impairs the regulation of protective Th17 cell response to intestinal and systemic Listeria monocytogenes (L. m) infection.9,11 Furthermore, P. UF1 regulates the neonatal T cells against necrotizing enterocolitis (NEC)-like injury in mice9 and enhances the neonatal protective T cells against intestinal pathogen infection over time.12 However, the bacterial effector mechanisms potentially instructing the function of colonic DCs to possibly control protective T cell immunity remain largely unknown. Here, we demonstrate that the glycosylation of bacterial LspA interacting with SIGNR1 is a pivotal factor, which transcriptionally and metabolically programs colonic DCs, leading to protective T cell activation in steady state and during intestinal infection. Further, glycosylated LspA-SIGNR1 interaction critically protects mice against colitis-induced intestinal barrier injury. Errors in the bacterial glycosylation significantly disrupt the intestinal homeostasis, manifesting in an inflammatory condition resulting in pathogen persistence and colonic tissue damage. Thus, this finding highlights the critical relevance of the glycosylated LspA in programming DC immunophysiology to mitigate pathogenic inflammation and the induced colitogenic potential in mice.
Results
Glycosylation of LspA by Pmt1
Knowing the significance of bacterial S-layer complexes in communicating with host cells,13 we sought to investigate the functional relevance of P. UF1 S-layer proteins potentially involved in the regulation of colonic DC function. One of the S-layer proteins of P. UF1 is LspA, which contains six N-terminal LGFP repeats [L-G-X-P-X(7-8)-D/N-G] involved in cell membrane anchoring and a C-terminal N-acetylglucosaminidase-like domain, potentially implicated in bacterial cell wall metabolism (Supplementary Fig. 1a). Phylogenetic analysis demonstrated that LspA was highly conserved in P. UF1 and closely related Propionibacterium strains. Moreover, LspA homologs were also found in evolutionarily distantly related bacterial species, including Bifidobacterium and Geodermatophilus (Supplementary Fig. 1b). Thus, to elucidate the functional significance of LspA within P. UF1 molecular machinery, the lspA gene was deleted from the bacterial chromosome, resulting in ΔlspA P. UF1 (Fig. 1a, b). ΔlspA P. UF1 demonstrated enhanced bacterial clusters and autoagglutination (Fig. 1c), suggesting the critical involvement of this protein in bacterial S-layer structures. Further, deletion of LspA significantly affected the bacterial transcriptomic and metabolomic signaling, including differential metabolic pathways involved in peptidoglycan biosynthesis, amino and nucleotide sugar metabolism, fructose and mannose metabolism (Supplementary Fig. 2a). The analyzed metabolites involved in protein glycosylation (e.g., GDP-mannose and mannose 1-phosphate), along with those important for cell wall metabolism (e.g., GlcNAc-6-phosphate and UDP-GlcNAc), were significantly deregulated within ΔlspA P. UF1 compared to P. UF1 (Supplementary Fig. 2b). RNA-Seq analysis further documented differentially expressed genes implicated in bacterial mannosylation and nucleotide sugar metabolism, including phosphatidylinositol mannosyltransferase pimA14 and GDP-mannose-dependent alpha-mannosyltransferase mgtA15 (Supplementary Fig. 2c). Thus, these data emphasize the importance of LspA in the regulation of glycan metabolism that may fundamentally impact the bacterial S-layer glycosylation.
Glycosylation of LspA by Pmt1. a Identification of ΔlspA P. UF1 strain. Genetic scheme for disruption of lspA gene by chromosomal insertion of plasmid pUCC-lspA (left). SDS-PAGE (middle) and Western blot (right) showing LspA protein was completely absent in ΔlspA P. UF1. CmR, chloramphenicol resistant gene. b Flow cytometric analysis of S-layer expression of LspA in P. UF1 and ΔlspA P. UF1 using anti-LspA serum antibodies. Control serum was derived from unimmunized mice. c Scanning electron microscopy (SEM) images of P. UF1 and ΔlspA P. UF1. SEM images in the bottom panel are magnified from the indicated zoom in the top panel. d ConA binding assay for S-layer proteins isolated from P. UF1 and ΔlspA P. UF1. e Neighbor-joining phylogenetic tree showing the relationship of Pmt proteins from Actinobacteria, Firmicutes, and Fungi. f qRT-PCR analysis of pmt1 expression in P. UF1 and Δpmt1 P. UF1. g SDS-PAGE analysis and ConA binding assay of S-layer proteins isolated from P. UF1, ΔlspA P. UF1, and Δpmt1 P. UF1. h SDS-PAGE analysis of purified glycosylated LspA (G-LspA) and non-glycosylated LspA (NG-LspA). i Equal amounts of purified G-LspA and NG-LspA proteins were separated by SDS-PAGE and analyzed by Western blot using anti-LspA antibodies, ConA binding assay, and ProQ Emerald 300 glycoprotein staining. Arrows indicate the LspA protein
The bacterial S-layer proteins are generally glycosylated for their noncovalent anchoring to the cell surface and interactions with environmental factors and host immune cells.5 Data demonstrated that the S-layer of P. UF1 reacted with concanavalin A (ConA), a mannose/glucose-binding lectin, while LspA deficiency resulted in the loss of ConA binding (Fig. 1d), suggesting that LspA may be glycosylated. Therefore, we investigated the glycosyltransferases responsible for adding glycol moieties to the bacterial S-layer using genome-wide bioinformatic analysis. Pmt1, a potential member of protein O-mannosyltransferase family responsible for mannose transfer to serine and threonine residues of proteins in yeast16, was identified in P. UF1 genome. Further analysis demonstrated that Pmt1 homologs fell into separate and loosely related groups of bacteria, including Actinobacteria (Fig. 1e). The pmt1 gene was then deleted in P. UF1 to assess the status of LspA glycosylation (Fig. 1f). Although Δpmt1 P. UF1 showed similar S-layer protein patterns when compared to P. UF1, no binding to ConA was observed for S-layer proteins isolated from Δpmt1 P. UF1 (Fig. 1g). To underscore the role of Pmt1 in the glycosylation of LspA, this protein was overproduced by ΔlspA P. UF1 and Δpmt1 P. UF1 strains to isolate and purify the glycosylated LspA (G-LspA) and non-glycosylated LspA (NG-LspA), respectively (Fig. 1h). While both G-LspA and NG-LspA were recognized by anti-LspA serum antibodies, only purified G-LspA bound to ConA and illuminated staining for glycoprotein (Fig. 1i). Thus, Pmt1 is critically required for the glycosylation of LspA.
O-mannosylated LspA interaction with SIGNR1
To elaborate on the nature of LspA glycosylation, the purified G-LspA and NG-LspA proteins were treated with PNGase F to release any N-glycans, permethylated and analyzed by MALDI-MS. Here, no N-linked glycans were detected in either of the LspA proteins (Supplementary Fig. 1c). The O-linked glycans were released by β-elimination procedure and permethylated prior to MALDI-MS analysis. Signals corresponding to Hex1-Hex6 were observed in the G-LspA protein (Fig. 2a), but not in the NG-LspA (Supplementary Fig. 1d). Furthermore, glycan compositional analysis demonstrated that mannose (Man) was the major monosaccharide of G-LspA, with a retention time of 10.9 min (Fig. 2b). In contrast, NG-LspA showed no traces of Man (Supplementary Fig. 1e). Note that a minor peak of glucose was also detected in both samples. However, glucose, as a very common contaminant, could be derived from reagents and detected as a free and minor glucose peak in HPAEC analysis. Moreover, glycomic analysis of released oligosaccharides demonstrated that Man3 was the major glycan in the G-LspA, comprising 77% of the total glycans. While Man2 and Man4 oligosaccharides were minor glycans, only traces of Man1, Man5 and Man6 were detected in the G-LspA (Fig. 2c).
Recognition of glycosylated LspA by SIGNR1. a O-linked glycan analysis of β-eliminated and permethylated G-LspA protein. Asterisk (*) indicates the contamination peak derived from reagents. b Glycosyl composition analysis of monosaccharides in the G-LspA sample. Trace levels of glucose were detected as a common contaminant derived from reagents. c Summary table showing the relative percentage of O-linked glycans from G-LspA. d LspA glycopeptides identified by glycoproteomics. The glycan composition and potential glycosylation sites (bolded) are shown. e Glycosyl linkage analysis of the O-glycans. Asterisks (*) indicate non-carbohydrate peaks. RT, retention time. f Binding of G-LspA to SIGNR1-hFc. Equal amounts of G-LspA and NG-LspA proteins were separated by SDS-PAGE and the specific interactions with SIGNR1-hFc were demonstrated, as no binding was detected in the presence of EDTA. g ELISA binding assays demonstrating G-LspA binding specificity with SIGNR1-hFc. The binding was abolished in the presence of EDTA, competitive zymosan, or blocking antibody to SIGNR1. h ELISA showing G-LspA did not bind to Dectin-1-hFc, as a control fusion protein. Zymosan served as a positive control. i Binding kinetics between G-LspA and SIGNR1-hFc. Various amounts of LspA proteins were coated on ELISA plates and incubated with SIGNR1-hFc (0.5 μg/ml). Binding was detected using HRP-conjugated anti-human IgG antibody. Kd, LspA concentration required to achieve a half-maximum binding with SIGNR1-hFc
The purified G-LspA protein was then digested with trypsin and elastase, resulting in peptides with >75% coverage (Supplementary Fig. 1a). LC-MS/MS analysis of the enriched peptides revealed seven O-glycopeptides at the N-terminus of LspA (Fig. 2d). In addition to the 41 threonine/serine residues involved in Man attachment, adjacent proline and alanine residues that may facilitate local conformational changes for protein O-glycosylation17 were also found in all the glycopeptides (Fig. 2d). Further, GC-MS analysis was performed to investigate the glycosyl linkages and positions of released O-glycans. Data demonstrated that Man oligosaccharides of LspA were short linear chains interconnected via (1 → 6)-linkage and (1 → 2)-linkage (Fig. 2e). Man(1 → 6)Man(1 → 2)Man was the major trisaccharide component in the G-LspA, while disaccharide Man(1 → 6)Man and tetrasaccharide Man(1 → 6)Man(1 → 2)Man(1 → 2)Man comprised a small percentage (Fig. 2e). These data indicate that LspA is a mannosylated S-layer glycoprotein with linear short-chain O-glycans.
SIGNR1 expressed by myeloid DCs recognizes characteristic molecular patterns with complex mannose and fucose structures in bacteria and fungi.18 Recently, we observed the binding of P. UF1 to SIGNR1, but not SIGNR3.9 To precisely delineate the role of G-LspA binding to SIGNR1 in regulating DCs to subsequently initiate T cell commitment, the interaction of G-LspA with SIGNR1 was biochemically investigated. Here, G-LspA and NG-LspA proteins were first separated by SDS-PAGE, transferred to PVDF membrane, and then incubated with SIGNR1-hFc fusion protein. This protein binding complex was analyzed by subsequent incubation with anti-human Fc secondary antibody. Data demonstrated that the purified G-LspA bound specifically to SIGNR1-hFc, and this binding was abolished in the presence of EDTA (Fig. 2f). Further, SIGNR1 interaction with G-LspA was assessed by ELISA showing G-LspA binding to SIGNR1-hFc (Fig. 2g), but not to Dectin-1-hFc used as a control fusion protein (Fig. 2h). This binding was completely blocked by pre-incubation of SIGNR1-hFc with anti-SIGNR1 antibody, or with the competitive ligand zymosan that is composed of β-glucan, α-mannan and mannosyl proteins (Fig. 2g). In contrast, no binding was observed for NG-LspA using similar assays (Fig. 2g). Furthermore, the G-LspA exhibited a dose-dependent binding with SIGNR1-hFc using protein concentrations ranging from 0.08 μg/ml to 10 μg/ml with a Kd value of 2.617 μg/ml G-LspA (Fig. 2i). In contrast, NG-LspA did not react with SIGNR1-hFc, even with higher protein concentrations up to 20 μg/ml (Fig. 2i), highlighting the specificity of SIGNR1 binding to glycosylated LspA.
Activation of colonic DCs by glycosylated LspA in steady state
SIGNR1 was majorly expressed by colonic CD11chi MHCIIhi DCs (Supplementary 3a, b). To shed light on the relevance of LspA glycosylation interacting with colonic DCs, CD11chi MHCIIhi CD11b+ F4/80− DCs were FACS sorted (Supplementary Fig. 4a, b) from mice gavaged with P. UF1 or ΔlspA P. UF1 to analyze their transcriptome by RNA-Seq. Data demonstrated the modulation of costimulatory molecules (Cd40, Cd80, Cd86, and Tnfsf4) in colonic DCs by P. UF1 (Fig. 3a). NF-κB signaling (Casp4, Traf1, Tnfrsf1b, Mapk6, Nfkbie, and Nfkbiz), cytokine/chemokine transcripts (e.g., Il1b, Il12b, Cxcl1, and Cxcl2), and antigen presentation-related genes (e.g., Serpinb9, Rab8b) were also significantly augmented in DCs derived from mice gavaged with P. UF1 compared to ΔlspA P. UF1. In contrast, DCs derived from ΔlspA P. UF1-gavaged mice showed activation of Sod3, Rhoh and Klf2, which may instruct functional suppression in these cells. Migrating DCs constitutively express genes with regulatory functions.19 Accordingly, DCs derived from mice gavaged with P. UF1 had elevated expression of genes, such as Cd274, Spred1, Etv3, Tnfnip2, Stat3, Stat4, and Stat5a. These cells were also enriched with genes implicated in DC development (Edn1, Cish), migration (Nrp2, Ccr10, Eps8), and differentiation (Pdk1, Hilpda), while DCs of ΔlspA P. UF1-gavaged mice exhibited increased quantities of genes suppressing cellular regulatory functions (e.g., Cyr61, Sdc1). In addition, transcription factor Irf4 controlling Th17 cell cytokine machinery,20 cell cycle inhibitor Cdkn1a involved in Treg cell formation,21 and T cell-attracting chemokines Ccl17 and Ccl22, were all significantly activated in DCs of P.UF1-gavaged mice. In contrast, ΔlspA P. UF1 enhanced the DC expression of Cd55 and Gilz genes associated with suppression of T cell function.22,23
Modulation of DC activation by glycosylated LspA in steady state. a–b C57BL/6 (Signr1+/+) mice were gavaged with P. UF1 or ΔlspA P.UF1, and colonic CD11chi MHCIIhi CD11b+ F4/80− DCs were isolated for RNA-Seq analysis. Heatmap showing a selection of top differentially expressed genes (FDR P < 0.05, fold change ≥ 1.5). Gene set enrichment analysis (GSEA) showing differentially enriched gene sets. Number signs (#) indicate gene sets without FDR correction (P < 0.05). c Colonic CD11chi MHCIIhi CD11b+ F4/80− DCs were FACS sorted from Signr1−/− and Signr1+/+ mice gavaged with P. UF1 or ΔlspA P. UF1 and transcripts of indicated genes in these groups of DCs were analyzed by qRT-PCR. Data are from 1 experiment (n = 3–4 mice/group). Error bars indicate SEM. *P < 0.05, **P < 0.01, ***P < 0.001, two-tailed unpaired t test
Gene set enrichment analysis (GSEA) demonstrated that gene sets for metabolic activities [e.g., glycolysis and oxidative phosphorylation (OXPHOS)] and DC activation, including activation of innate responses, regulation of I-κB/NF-κB signaling, cytokine receptor activity and cell migration, were enriched in DCs derived from mice gavaged with P. UF1 (Fig. 3b). In contrast, glycosylated LspA deficiency abrogated cellular protein responses in DCs, resulting in enhanced endoplasmic reticulum (ER) stress-associated activities (Fig. 3b), which were associated with increased transcripts of unfolded protein response (UPR)-related genes Xbp1, Derl3, and Edem2 in these cells (Fig. 3a). Further, P. UF1-induced DC activation (e.g., Cd80, Cd86, Il1b, and Il12b) was abrogated due to Signr1 deficiency (Fig. 3c). Moreover, loss of LspA glycosylation (ΔlspA P. UF1-gavaged Signr1+/+ mice) mirrored the deficiency of SIGNR1 (P. UF1-gavaged Signr1−/− mice), suggesting a role of glycosylated LspA-SIGNR1 interaction in DC activation.
Having shown that LspA deficiency impacted the transcriptome of DCs, we asked whether LspA deficiency would also influence the regulation of T cell polarization in steady state. Thus, C57BL/6 mice were gavaged with P. UF1 or ΔlspA P. UF1 to analyze colonic DC and T cell responses. Data demonstrated that ΔlspA P. UF1 compared to P. UF1 significantly decreased DC and T cell responses, including IL-10+ Th17 cells and 10+ Tregs (Supplementary Fig. 5a, b). Notably, no difference in intestinal bacterial colonization was observed in mice gavaged with P. UF1 or ΔlspA P. UF1, as both strains transiently colonized conventional mice (Supplementary Fig. 5c), suggesting that glycosylated LspA is not involved in bacterial colonization, but instructs DC activation to polarize T cells.
Programming transcriptomic and metabolomic DC-machineries during intestinal infection
Intestinal DCs rapidly respond to invading pathogens.24 However, these cells may be functionally conditioned by pathological surrounding signals induced by intestinal infections.25 To elaborate on the bacterial glycosylated LspA influencing DC transcriptome, colonic MHCIIhi CD11chi CD11b+ F4/80− DCs were FACS sorted from mice gavaged with P. UF1 or ΔlspA P. UF1 and then orally infected with ΔactA L. m. RNA-Seq analysis demonstrated that P. UF1 controlled the expression of genes associated with DC activation (e.g., Cstl, Tsc1) and suppression of TGF-β signaling (e.g., Tgbfi, Eaf2, and Cited2) (Fig. 4a). Further, transcripts of Tnip3 regulating NF-κB activation and Dok-2 suppressing Ras-Erk signaling were stimulated in colonic DCs isolated from mice gavaged with P. UF1. Importantly, Cd209b (Signr1), whose activation highly depends on the mannosylation of glycans, was upregulated in DCs derived from mice gavaged with P. UF1. In contrast, Toll-like receptors (Tlr3, Tlr4, and Tlr12) associated with proinflammatory response were upregulated in DCs of mice gavaged with ΔlspA P. UF1 (Fig. 4a, b). Mx1 and Irf1, selectively activated during TLR-induced DC stimulation,26 were also enriched in DCs derived from ΔlspA P. UF1-gavaged mice. Consistently, a proinflammatory status was readily observed in DCs of mice gavaged with ΔlspA P. UF1. Accordingly, a set of proinflammatory genes (e.g., Ifng, Tnfsf15, and Ifi204), several genes associated with cell apoptosis (e.g., Daxx, Nab2, and Fbxw7), and stress-associated activities were activated in DCs derived from mice gavaged with ΔlspA P. UF1 (Fig. 4a, b). In addition, GSEA demonstrated enhanced mitochondrial biogenesis in DCs isolated from P. UF1-gavaged mice, as indicated by enriched gene sets for OXPHOS, TCA cycle and respiratory chain, and NADH dehydrogenase complex (Fig. 4c). Moreover, pathways regulating apoptosis and protein localization to ER were also enhanced in these cells. In contrast, DCs from ΔlspA P. UF1-gavaged mice exhibited dysregulated protein folding and ER stress-associated activities (Fig. 4c), consistent with increased transcripts of the UPR-related genes Xbp1 and Derl3 in these cells (Fig. 4a, b).
Transcriptomic programming of colonic DCs by P. UF1 expressing glycosylated LpsA during Listeria infection. C57BL/6 mice were gavaged with P. UF1 or ΔlspA P. UF1 and then orally infected with ΔactA L. m. Colonic CD11chi MHCIIhi CD11b+ F4/80− DCs were FACS sorted for transcriptomic analysis seven days after infection. a Heatmap showing significantly differentially expressed genes (FDR P < 0.05, fold change ≥ 1.5) in DCs derived from mice gavaged with P. UF1 compared to ΔlspA P. UF1. b qRT-PCR analysis of a selection of differentially expressed genes. Results are presented as fold change over DCs of P. UF1-gavaged mice. c GSEA showing differentially enriched pathways. Number signs (#) indicate gene sets without FDR correction (P < 0.05). Data are from 1 experiment (n = 4 mice/group). Error bars indicate SEM. *P < 0.05, **P < 0.01, two-tailed unpaired t test
Inflammatory DC response is associated with declined mitochondrial activity promoted by TLR signaling.27 Having demonstrated that glycosylated LspA controlled TLR-activation and enriched gene sets for OXPHOS (Fig. 4), we further assessed whether P. UF1 expressing glycosylated LspA would maintain mitochondrial respiration during intestinal infection. Thus, colonic DCs were enriched by magnetic beads from mice gavaged with P. UF1 or ΔlspA P. UF1 and orally infected with ΔactA L.m to analyze the real-time changes in the rate of extracellular acidification (ECAR), a measurement of glycolysis, and the mitochondrial rate of oxygen consumption (OCR). While no difference in the ECAR was observed, OCR was significantly decreased in enriched DCs derived from mice gavaged with ΔlspA P. UF1 compared to DCs derived from P. UF1-gavaged mice (Fig. 5a). These data support the notion that glycosylated LspA may regulate DC metabolic function during intestinal infection.
Metabolomic programming of colonic DCs by P. UF1 expressing glycosylated LspA during Listeria infection. a Colonic DCs were enriched from mice gavaged with P. UF1 or ΔlspA P. UF1 and orally infected with ΔactA L. m. The mitochondrial respiration of enriched DCs by magnetic beads was evaluated by measuring their real-time changes in the ECAR and OCR in response to sequential addition of oligomycin (Oligo), FCCP, rotenone (Rot), and antimycin A (Ant). Bar graphs show the basal ECAR and OCR. b Colonic CD11chi MHCIIhi CD11b+ F4/80− DCs were FACS sorted from mice gavaged with P. UF1 or ΔlspA P. UF1 and orally infected with ΔactA L. m. The cellular metabolome of DCs was analyzed by high-resolution mass spectrometry. Heatmap showing the distinctive metabolomes of colonic DCs. c Metabolic pathway analysis of metabolites with intensities significantly altered by comparing FACS sorted DCs derived from P. UF1- and ΔlspA P. UF1-gavaged mice. Dashed line shows the permutation P value of 0.05. d Scatter plots for selected metabolite features, with putative annotation, in the significant pathways identified by Mummichog. The m/z, retention time (in seconds) and adduct ion were labeled for each metabolite. Data are from 1 experiment (n = 4–5 samples/group). Error bars indicate SEM. *P < 0.05, **P < 0.01, ***P < 0.001, two-tailed unpaired t test
It was recently demonstrated that DC function can be impacted by cellular metabolic factors that facilitate the biosynthetic and bioenergetic needs of these cells.28 To elaborate on this notion, colonic MHCIIhi CD11chi CD11b+ F4/80− DCs were FACS sorted from mice gavaged with P. UF1 or ΔlspA P. UF1 and orally infected with ΔactA L. m to analyze the metabolomic activity of these cells. Here, a distinct metabolome was identified in DCs derived from P. UF1-gavaged mice compared to those derived from ΔlspA P. UF1-gavaged mice (Fig. 5b). Metabolic pathways, including arginine and proline metabolism, purine metabolism and de novo fatty acid biosynthesis, were differentially activated in DCs isolated from the aforementioned groups of mice (Fig. 5c). Putatively annotated eicosatrienoic acid, known as an anti-inflammatory metabolite, 4-aminobutanal indicative of anti-inflammatory putrescine metabolism, and proline involved in suppressing reactive oxygen species (ROS),29 were markedly accumulated in DCs from P. UF1-gavaged mice compared to the other group (Fig. 5d). In contrast, putative metabolites associated with proinflammation and energy starvation [e.g., methylimidazoleacetic acid and 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR)] and cell apoptosis-inducing deoxyadenosine were significantly enriched in DCs derived from ΔlspA P. UF1-gavaged mice (Fig. 5d). Thus, these data specifically demonstrate the requirement of glycosylated LspA expressed by P. UF1 that programs the regulation of DC function during Listeria-induced inflammation.
Regulating protective T cell response by P. UF1 expressing glycosylated LspA
To further elucidate the functional relevance of glycosylated LspA in modulating colonic homeostasis during intestinal infection, C57BL/6 mice were gavaged with P. UF1, ΔlspA P. UF1 or PBS, and then orally infected with ΔactA L. m. Notably, P. UF1, compared to ΔlspA P. UF1, regulated proinflammatory responses by controlling the frequencies and cell counts of IL-1β+, IL-6+, and IL-12/23p40+ DCs (Fig. 6a). However, such induced DC regulation was abolished in Signr1−/− mice gavaged with P. UF1, ΔlspA P. UF1 or PBS and subsequently infected with ΔactA L. m (Fig. 6a), denoting a potential role of glycosylated LspA-SIGNR1 interaction in controlling inflammatory DC response. Further, tuned DC response was associated with T cell regulation, leading to an increased percentage and number of Th17 and IL-10+ Treg cells in mice gavaged with P. UF1 compared to ΔlspA P. UF1 or PBS (Fig. 6b). Loss of protective immunity in Signr1+/+ mice gavaged with ΔlspA P. UF1 resulted in delayed ΔactA L. m clearance compared to mice gavaged with P. UF1 (Fig. 6c). In addition, transcripts of proinflammatory molecules such as Il6, Il12b, Ifng, Ccl5, and Cxcl2 were significantly decreased in colonic tissues of Signr1+/+ mice gavaged with P. UF1 compared to those gavaged with ΔlspA P. UF1 or PBS (Fig. 6d). In contrast, the protective T cell immunity was blunted in Signr1−/− mice gavaged with P. UF1, ΔlspA P. UF1 or PBS and then infected with ΔactA L. m (Fig. 6b), and no difference in pathogen clearance was observed in these groups of mice (Fig. 6c). Furthermore, data demonstrated that deficiency in bacterial glycosylated LspA did not induce any changes in the gut microbial composition of mice gavaged with ΔlspA P. UF1 compared to P. UF1 during ΔactA L. m infection (Supplementary Fig. 6a, b). Together, these results indicate the crucial role of glycosylated LspA-SIGNR1 interaction in regulating colonic DCs to orchestrate protective T cell response to intestinal Listeria infection.
Requirement of glycosylated LspA for protective T cell regulation. Signr1+/+ and Signr1−/− mice were gavaged with P. UF1, ΔlspA P. UF1 or PBS and then orally infected with ΔactA L.m. Seven days post infection, colonic DC and T cell responses were analyzed. a Percentages and total counts of IL-1β+, IL-6+, and IL-12/23p40+ DCs in Signr1+/+ (top panel) and Signr1−/− mice (bottom panel). Flow plots are from Signr1+/+ mice. b Percentages and total counts of Th17, IL-10+ Th17, and IL-10+ FoxP3+ Treg cells in Signr1+/+ (top panel) and Signr1−/− mice (bottom panel). Flow plots are from Signr1+/+ mice. c Box and whisker plots of fecal ΔactA L. m burdens measured in Signr1+/+ (left panel) and Signr1−/− mice (right panel) on days 1–4 after infection. Dashed lines represent the limit of pathogen detection. d Box and whisker plot showing transcript levels of proinflammatory molecules in the colonic tissues of Signr1+/+ mice. Data are representative of 2 (Signr1−/− mice) or 3 (Signr1+/+ mice) independent experiments (n = 5 mice/group, a, b) or pooled from 2 independent experiments (n = 9–10 mice/group, c, d). Error bars indicate SEM. *P < 0.05, **P < 0.01, ANOVA plus Tukey’s post-test (a, b) or Kruskal-Wallis plus Dunn’s post-test (c, d)
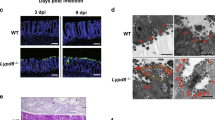
Ameliorating dextran sulfate sodium (DSS)-induced colitis by P. UF1 expressing glycosylated LspA
IL-17A and IL-10+ Tregs contribute to protection against acute intestinal colitis.30,31,32 To further investigate the protective role of glycosylated LspA expressed by P. UF1 in chemically induced colitis, groups of mice were gavaged with P. UF1, ΔlspA P. UF1 or PBS, and the experimental colonic tissue damage was induced by 3% DSS. Data demonstrated that P. UF1, compared to other groups of mice, significantly reduced the disease severity, as indicated by the reduction of weight loss, diarrhea and fecal blood scores (Fig. 7a, b), and increased colon length (Fig. 7c). Furthermore, severe signs of inflammation-induced thickening of the bowel wall and loss of regular blood vessel structures were observed in the colons of PBS- and ΔlspA P. UF1-gavaged mice but not in P. UF1-gavaged mice (Fig. 7d). Histological analysis demonstrated severe tissue destruction, characterized by extensive segmental to diffuse mucosal epithelium and crypt loss with associated mucosal and submucosal inflammation in the colonic tissues of mice gavaged with PBS or ΔlspA P. UF1, while DSS-induced colitis was substantially mitigated in mice gavaged with P. UF1 (Fig. 7e, f). Furthermore, obtained data also demonstrated that DSS-treated mice gavaged with P. UF1 were markedly protected from leaky gut when compared with other groups, as documented by FITC-dextran assay (Fig. 7g). Consistently, enhanced transcripts of tight junction proteins (Cldn2, Cldn3, Cldn7, Cldn8, and ZO-3) in colonic tissues of P. UF1-gavaged mice were observed compared to the other groups (Fig. 6h). Thus, glycosylated LspA expressed by P. UF1 contributes to the maintenance of intestinal barrier integrity.
Contribution of glycosylated LspA-SIGNR1 interaction to protection against DSS-induced colitis. Signr1+/+ and Signr1−/− mice were gavaged with P. UF1, ΔlspA P. UF1 or PBS on days −7, −4, and −1, and 3% DSS was given in drinking water on day 0. Mice continued to receive the bacteria every three days for an additional three times and monitored for disease progression until day 12 (Signr1+/+ mice) or day 10 (Signr1−/− mice due to disease severity). a Weight loss, diarrhea, and fecal blood scores were monitored over time. b Bar graphs showing area under curve (AUC) for diarrhea and FOB scores. c Colon lengths were measured and compared. d Colonoscopies were performed in the indicated groups of mice. e–f Colitis scores based on histopathology of the colons were also used as measures of disease severity. g Serum FITC-dextran levels of DSS-treated Signr1+/+ and Signr1−/− mice gavaged with P. UF1, ΔlspA P. UF1 or PBS. h–i Transcript levels of tight junction proteins in colonic tissues of the indicated groups of mice. Data are pooled from 2 independent experiments (n = 7–8 mice/group). Error bars indicate SEM. **P < 0.01, ***P < 0.001, ****P < 0.0001, ANOVA plus Tukey’s post-test
To further elucidate the protective role of glycosylated LspA-SIGNR1 signaling in DSS-induced colitis, Signr1−/− mice were gavaged with P. UF1, ΔlspA P. UF1 or PBS and treated with DSS. Once again, lack of SIGNR1 interaction with glycosylated LspA resulted in no improvement of colitogenic disease progression in any of the groups of mice (Fig. 7a–f). Here, the disruption of glycosylated LspA-SIGNR1 interaction abolished the regulation of intestinal permeability (Fig. 7g) and blunted the expression of tight junction proteins in Signr1−/− mice (Fig. 7i). Collectively, these data suggest that glycosylated LspA interacting with SIGNR1 sustains gut homeostasis to protect against detrimental signals inducing tissue damage.
Discussion
The maintenance of gut homeostasis requires a rigidly balanced dialog between the resident microbes and the host.1 This can be established through the interaction of a variety of bacterial gene products with their cognate receptors expressed by phagocytic cells.6 Disruption of such elegantly induced communication may result in pathogenic inflammation and intestinal tissue damage.33 In this study, we demonstrate a protective mechanism at the intestinal interface whereupon glycosylated LspA interacting with SIGNR1 dictates DC response that in turn regulates protective T cells against intestinal pathogen infection and chemically induced colitis.
There has recently been a growing interest in the bacterial S-layer proteins with multiple regulatory properties.34,35 Accordingly, the composition of the S-layer proteins of Propionibacterium species is remarkably variable, and the functions of bacterial S-layer proteins are strain-dependent.36 For instance, surface layer protein B (SlpB), highly expressed by some of P. freudenreichii strains, facilitates bacterial adhesion to epithelial cells.37 While no SlpB homolog is found in P. UF1, LspA constitutes the major extractable S-layer protein in this bacterium. The high protein expression may also suggest that LspA, particularly in P. UF1, is a valuable factor for not only supporting the S-layer structure of this bacterium but could also be important for fine-tuning intestinal immunity and may serve as an elegant vehicle for mucosal vaccine and therapeutic approaches.38 Although LspA is conserved, the levels of protein expression vary dramatically in different Propionibacterium strains,35 indicating potentially differential gene regulation in these bacterial strains. With this notion in mind and the unique expression pattern of LspA by P. UF1, we were prompted to further elaborate on its physiological characteristics and its potential implication in regulating intestinal immunity. Here, we demonstrate that deletion of lspA leads to fundamental changes in pathways associated with S-layer carbohydrate metabolisms, including mannosylation, suggesting the critical role of LspA in maintaining the S-layer glycosylation profile. Indeed, ConA binding assays comparing S-layer proteins isolated from P. UF1 and ΔlspA P. UF1 show that glycosylation of other potential glycoproteins is also likely impaired by LspA deficiency. Further, changes in S-layer glycosylation may directly impact the bacterial cell-cell interactions, leading to enhanced autoagglutination due to LspA deficiency in P. UF1, possibly as a result of altered cell surface hydrophobicity. This is consistent with the observations that surface glycosylation, such as flagella glycosylation, is highly associated with autoagglutination, which is an important step for microcolony formation on the intestinal epithelial cells that contributes to intestinal colonization.39,40
In this study, we also clearly demonstrate that LspA is an O-mannosylated glycoprotein, uniquely representing the first glycoprotein characterized in Propionibacteria. Furthermore, the glycosylation of LspA critically requires Pmt1 activity. This feature is reminiscent of protein glycosylation in Streptomyces and Mycobacterium, whereupon the O-glycosylation of surface lipoproteins or phosphate-binding protein PstS, are highly dependent on the membrane-associated lipoprotein Pmt.41 Thus, the Pmt-mediated protein O-mannosylation seems to be a general pathway in actinomycetes. In fungi and yeasts, up to seven Pmt family members have been identified thus far, and the homomeric and/or heteromeric interactions among Pmt members cooperatively initiate the protein glycosylation, resulting in the biosynthesis of diversified glycan structures.42 However, Pmt1 seems to be the only protein glycosyltransferase in P. UF1, and no other protein glycosylation is observed in the S-layer proteins isolated from Δpmt1 P. UF1. Furthermore, the linear short-chain mannose is found to be the major glycan structure of bacterial LspA; thus, concluding that Pmt1 is the crucial enzyme responsible for modifying S-layer proteins, particularly LspA, with simple mannoses in P. UF1.
Recognition of microbial gene products by sensing receptors, including C-type lectins, is essential for translating the nature of microbes into gene-transcriptional and metabolic programs that may initiate the regulation of DC function to prime T cell polarization.43 SIGNR1, a murine homolog of human DC-SIGN, conditions intestinal DCs for the induction of oral tolerance44 and plays a key role in host defense against pathogen infection.45 However, further rigid molecular studies are still required to mechanistically elucidate the gene and metabolic programs that are modulated by the interactions of this sensing receptor with the bacterial surface glycosylation. Here, we demonstrate that LspA glycosylation is required to be recognized by its receptor, SIGNR1, leading to the regulation of DC activation in steady state and during pathogen-induced inflammation. Accordingly, in steady state P. UF1, via its glycosylated LspA, seems to optimally induce the activation of colonic DCs when compared with its counterpart, ΔlspA P. UF1. Mainly, P. UF1 regulates the expression of costimulatory, cytokine and antigen presentation molecules (e.g., Cd40, Il1b, Serpinb9, Rab8b, and Bcl3) in colonic DCs while glycosylated LspA deficiency results in the activation of suppressive molecules (e.g., Sod3, Rhoh and Klf2, Cyr61, Sdc1) that may dysfunction colonic DCs to properly induce T cell differentiation. More importantly, P. UF1 decorated with glycosylated LspA regulates DCs that control T cell response to pathogen infections. Here, regulatory signaling appears to be activated by LspA glycosylation, as Tgfbi, Eaf2, and Cited2 genes associated with suppression of TGF-β signaling are tuned down in DCs derived from mice gavaged with P. UF1 compared to ΔlspA P. UF1. Further, proinflammatory genes (e.g., Ifng and Ifi204) are also downregulated in DCs derived from mice gavaged with P. UF1. Consistently, anti-inflammatory polyunsaturated fatty acid eicosatrienoic acid is highly enriched in DCs of P. UF1-gavaged mice, while proinflammatory metabolites, such as methylimidazoleacetic acid and AICAR, are significantly elevated in DCs of ΔlspA P. UF1-gavaged mice during Listeria infection. In addition, DCs derived from ΔlspA P. UF1-gavaged mice exhibit significant quantities of genes associated with cell apoptosis and stress response, consistent with decreased cellular proline levels that are important for protecting against various cellular stresses.29 This may emphasize the impaired DC metabolism induced by glycosylated LspA deficiency, which is further supported by reduced mitochondrial respiration and enriched metabolite AICAR that serves as a metabolic activator responding to energy starvation of the cells. Collectively, these data suggest that LspA glycosylation may be an important factor that modulates colonic DC response via transcriptomic and metabolic reprogramming.
Proinflammatory DC regulation may impact T cell polarization.7 Indeed, dysregulated cellular pathways in DCs induced by ΔlspA P. UF1 significantly reduce the protective T cell immunity and correspondingly manifest in uncontrolled intestinal pathogen infection, resulting in delayed pathogen clearance and enhanced intestinal inflammation. In addition, no protection is seen in mice when either SIGNR1 or its ligand, glycosylated LspA, is deficient, indicating the critical role of the glycosylated LspA-SIGNR1 axis in regulating colonic DC response that dictates protective T cell response against pathogen infection. We have previously shown that P. UF1 induces DlaT-specific Th17 cells, which are indispensable for protection against Listeria infection.9,11,12 While DlaT expression is not impacted by lspA deletion (Supplementary Fig. 2c), induced Th17 cells notably require a regulated cytokine environment initiated by DCs through glycosylated LspA interacting with SIGNR1.
Interestingly, glycosylated LspA deficiency does not impact the phylum of gut microbiota in bacterially-gavaged groups of mice and then infected with Listeria, suggesting that glycosylated LspA may not contribute to any changes in the intestinal bacterial community. Yet, do changes in bacterial metabolome due to glycosylated LspA deficiency contribute to reduced regulation of DCs during intestinal infection? Although critical metabolic changes are observed within ΔlspA P. UF1, we posit that these metabolites may be mostly restricted to intracellular bacterial metabolic networks in response to altered S-layer glycosylation and may not directly impact the immune cells. Nonetheless, this important notion still requires further rigid investigations.
Ample data demonstrate that IL-17A serves as a protective factor against DSS-induced colitis by maintaining intestinal tight junctions and promoting epithelial repair.31,46 Here, Th17 cells, together with IL-10+ Tregs, regulated by colonic SIGNR1+ DCs interacting with glycosylated LspA, may be required for ameliorating DSS-induced colitis, all of which potentially result in the maintenance of tight junction expression that controls the intestinal permeability and the mitigation of chemically induced inflammation and intestinal tissue damage.
In summary, our data demonstrate the molecular mechanism through which DC functions can be fine-tuned via bacterial glycosylated LspA interacting with SIGNR1. Such a regulated DC response is pivotal in priming protective T cell response to intestinal infection and plays a critical role in mitigating DSS-induced colitis in mice. Induced immune regulatory processes involving the fine-tuned receptor-ligand interaction, mainly glycosylated LspA-SIGNR1, elicits functional gut homeostasis during the intestinal inflammatory condition. Thus, consistent with this finding, glycosylation of a bacterial gene product such as LspA may be an important feature for a future formulated prebiotic, conjugated vaccines and therapeutic targets that not only induce the regulation of innate and protective T cells against intestinal pathogen challenges, but may also prevent induced pathogenic inflammation that triggers tissue damage and the progression of intestinal proinflammatory diseases (e.g., colitis) in affected patients.47,48 Thus, a deeper understanding of beneficial bacterial gene products influencing the regulation of gut homeostasis may pave the way for the development of new pre or postbiotic therapeutic strategies to potentially treat inflammatory bowel disease (IBD) or colon cancer.49,50 Finally, our work uniquely illustrates the significance of the mannosylated ligand, bacterial LspA, and its critical binding to SIGNR1 that sustains the optimal activation and regulation of colonic DCs in intestinal steady state and during inflammatory condition. Conclusively, shedding light on the relevance of a sensing receptor, SIGNR1 and its human homolog, DC-SIGN, may reveal the critical innate factor in resisting detrimental signals inducing tissue damage that manifest in IBD. Thus, activating critical signals involving SIGNR1 (DC-SIGN) in health and human diseases may advance our vision to improve and develop therapeutic platforms mitigating IBD, which increasingly affects more than 3.5 million worldwide.51
Materials and methods
Mice
C57BL/6 mice (6–9 weeks old) were obtained from Jackson Laboratory and maintained under specific pathogen-free, Helicobacter-free conditions. Signr1−/− mice were provided by Huang Shau-Ku (Johns Hopkins University School of Medicine, Baltimore, Maryland, USA). All animal studies were approved by the Animal Care and Use Committee of the University of Florida under the protocol number 201708484. Mice were maintained in accordance with the Animal Welfare Act and the Public Health Policy on Humane Care.
Bacterial administration
In steady state, Signr1+/+ and Signr1−/− mice were gavaged with P. UF1 or ΔlspA P. UF1 (109 CFU/mouse/100 μl) every 3 days over the course of 12 days, and mice were euthanized on day 14 to isolate colonic immune cells. During ΔactA L. m infection, Signr1+/+ and Signr1−/− mice were gavaged with P. UF1, ΔlspA P. UF1 (109 CFU/mouse/100 μl) or PBS on days −7, −4, −1, and 2. Mice were denied food for 4 h on day 0 and then orally infected with 100 μl PBS containing 50 mg/ml CaCO3 and approximately 109 CFU of ΔactA L. m. Fecal samples were collected on days 1–4, and mice were sacrificed on day 7. Pathogen loads were determined by plating serial dilutions of fecal samples on BHI agar supplemented with 200 μg/ml streptomycin.
Cell isolation and flow cytometry
Density gradient centrifugation using Percoll was performed to isolate lamina propria cells from mouse colon. Briefly, tissues were collected and fecal contents in the colon were carefully removed. Colons were opened longitudinally and cleaned with cold PBS and then shaken in PBS containing 20 mM Hepes and 10 mM EDTA for 30 min at 37 °C. Tissues were cut into small pieces and incubated with digestion solution [RPMI 1640 containing 10% FBS (Thermo Fisher Scientific, Waltham, MA), 0.4% β-mercaptoethanol, 400 U/ml collagenase VIII (Sigma Aldrich, St. Louis, MO) and 100 μg/ml DNase I (Sigma Aldrich, St. Louis, MO)] for 1.5 h at 37 °C in a 5% CO2 incubator. Digested tissues were filtered through 100 μm cell strainer (Genesee Scientific, San Diego, CA), and cells were resuspended in 5 ml of 40% Percoll (Sigma Aldrich, St. Louis, MO) and overlaid on 5 ml of 80% Percoll. Cells in the interphase were collected after gradient centrifugation (1258 g, 25 min, 25 °C).
Flow cytometry was performed as described previously9 with some modifications. Isolated cells were stimulated with 50 ng/ml PMA (Sigma Aldrich, St. Louis, MO) and 500 ng/ml ionomycin calcium salt (Sigma Aldrich, St. Louis, MO) for 4 h and 5 μg/ml brefeldin A (BioLegend, San Diego, CA) was added 2 h before cells were harvested. Stimulation was performed in IMDM medium (Sigma Aldrich, St. Louis, MO) containing 10% FBS, 1% penicillin/streptomycin, 0.4% β-mercaptoethanol, 10 mM Hepes and 2 mM L-Glutamine. Dead cells were detected using LIVE/DEAD® Fixable Blue Dead Cell Stain Kit (Thermo Fisher Scientific, Waltham, MA), followed by incubation with Mouse Fc Blocking Reagent (Miltenyi Biotec, Auburn, CA). Cells were first stained for cell-surface markers and then resuspended in fixation/permeabilization solution [Cytofix/Cytoperm kit (BD Biosciences, San Diego, CA) for DC cytokine analysis, or Foxp3 Transcription Factor Staining Buffer Set (eBiosciences, San Diego, CA) for T cell analysis]. Cells were stained with following fluorescent antibodies: eVolve 655-CD45 (catalog 86-0451-42), eFlour506-CD11c (catalog 69-0114-80), PE-MHCII (catalog 12-5321-82), PE/Cy5-F4/80 (catalog 15-4801-80), APC-SIGNR1 (catalog 17-2093-82), PE/Cy5-CD8 (catalog 15-0081-83), PE/Cy7-Pro-IL-1β (catalog 25-7114-82)/rat IgG1 κ, FITC-IL-6 (catalog 11-7061-82)/rat IgG1 κ, eFlour 450-IL-12/23p40 (catalog 48-7123-82)/mouse IgG1 κ, and eFlour450-FoxP3 (catalog 48-5773-82)/rat IgG2a κ from Thermo Fisher Scientific; APC/Cy7-CD11b (catalog 101226), PerCP/Cy5.5-CD64 (catalog 139308), APC/Cy7-CD3 (catalog 100330), Brilliant Violet 605-CD4 (catalog 100548), PE-IL-17A (catalog 506904)/rat IgG1 κ, and FITC-IL-10 (catalog 505006)/rat IgG2b κ from BioLegend. Data were collected by an LSR II Fortessa (BD Biosciences, San Jose, CA) and analyzed with FlowJo software (version 10) (TreeStar, Ashland, OR). After dead and doublet cell exclusion, and the subsequent CD45+ selection, DCs were defined as CD11chi MHCIIhi CD11b+ F4/80− (Supplementary Fig. 7a) and T cells as CD3+ CD8− CD4+ (Supplementary Fig. 7b).
DC sorting
Colonic cell suspensions were prepared from mice gavaged with P. UF1 or ΔlspA P. UF1 in steady state or during intestinal ΔactA L. m infection. Cells were labeled with a cocktail of fluorescent antibodies specific for: PE/Cy7-CD11c (BioLegend, catalog 117318), PE-MHCII (Thermo Fisher Scientific, catalog 12-5321-82), APC-CD11b (BioLegend, catalog 101212), FITC-F4/80 (Thermo Fisher Scientific, catalog 11-4801-82). Dead cells were identified and excluded using LIVE/DEAD Fixable Violet Dead Cell Stain (Thermo Fisher Scientific, Waltham, MA). CD11chi MHCIIhi CD11b+ F4/80− DCs (Supplementary Fig. 4a) were isolated using a SONY SH800S Cell Sorter (Sony, Tokyo, Japan). The purity of sorted cells analyzed by flow cytometry was determined to be >98% (Supplementary 4b). Subsequently, cell pellets were resuspended in RLT plus buffer (Qiagen, Germantown, MD) for RNA extraction or snap-frozen in liquid nitrogen for metabolomic analysis.
RNA-Seq analysis
Total RNA was extracted from about 1 × 104 CD11chi MHCIIhi CD11b+ F4/80− DCs isolated from each individual mouse using an RNeasy Plus Micro Kit (Qiagen, Germantown, MD). cDNA was generated using a SMART-Seq HT kit (Takara Bio Inc., Mountain View, CA) and RNA-Seq libraries were constructed using a Nextera XT DNA Library Preparation Kit (Illumina, Inc., San Diego, CA). Barcoded samples were sequenced on an Illumina HiSeq instrument (Illumina, Inc., San Diego, CA) at the University of Florida ICBR NextGen DNA Sequencing Core Facility. The sequencing reads were mapped to the Mus musculus genome (NCBI GRCm38/mm10) using STAR aligner (v2.6.0), and count table was generated using SubReads featureCounts (v1.6.0). Significantly altered genes (RPKM ≥ 1, FDR P < 0.05, fold change ≥ 1.5) were identified by DESeq2. Regularized-log-transformation of count data was performed for heatmap plotting. Gene set enrichment analysis (GSEA) was performed in the javaGSEA (v3.0) using GO, Hallmark, KEGG and REACTOME database.
Extracellular flux analysis
Colonic DCs were isolated from mice gavaged with P. UF or ΔlspA P. UF1 and orally infected with ΔactA L. m using a Pan-DC Enrichment Kit (StemCell Technologies, Vancouver, Canada). For real-time analysis of extracellular acidification rate (ECAR) and oxygen consumption rate (OCR), DCs were analyzed using an XFe96 Extracellular Flux Analyzer (Seahorse Bioscience, North Billerica, MA) as described.52 Briefly, enriched colonic DCs (3 × 105 cells/well, pooled from 3 mice) were analyzed in non-buffered RPMI medium supplemented with 2.5 μM dextrose, 2 nM glutamine, and 1 μM sodium pyruvate. ECAR and OCR were analyzed in response to 1 μM oligomycin, 1.25 μM fluoro-carbonyl cyanide phenylhydrazone (FCCP), 1 μM rotenone and 1 μM antimycin A.
High-resolution metabolomics analysis
FACS sorted colonic CD11chi MHCIIhi CD11b+ F4/80− DCs were isolated from mice gavaged with P. UF1 or ΔlspA P. UF1 and orally infected with ΔactA L. m. Note: colonic cells of 2 mice were combined to obtain 60,000–80,000 DCs/sample. After vortex and incubation with acetonitrile-water (2:1) at 4 °C for 30 min, DC samples were centrifuged and the supernatants were analyzed by LC-MS. Each sample was run in triplicates on an Orbitrap Fusion Tribrid Mass spectrometer with the resolution of 120,000 (Thermo Fisher, San Diego, CA), with dual chromatography using a 5 min C18 reversed-phase chromatography in negative electrospray ionization (ESI) mode and a 5 min HILIC chromatography in positive ESI mode over a mass-to-charge ratio (m/z) range of 85–1250. Student’s t-test was performed between treatment groups. Subsequently, metabolic pathway analysis was performed by Mummichog software (v2.0) with default parameters. 786 significant metabolite features in negative mode and 1234 metabolite features in positive mode were used as input to Mummichog. The pathways represented by at least two significant metabolites and enriched at P < 0.05 in positive mode (HILIC column) are presented.
DSS-induced colitis
Signr1+/+ and Signr1−/− mice were treated with 3% DSS in drinking water for 5 consecutive days (made fresh every 2–3 days) to induce colitis. Mice were monitored for disease progression through day 12 (Signr1+/+ mice) or day 10 (Signr1−/− mice as they are more susceptible to induced colitis) after DSS treatment. For bacterial administration, mice were orally gavaged with P. UF1, ΔlspA P. UF1 or PBS on days −7, −4, −1, 2, 5, and 8 for a total of 6 gavages. Colitis severity was determined by histopathology. Tissues were fixed, sectioned, and stained with hematoxylin and eosin (Histology Tech Services, Gainesville, FL). Stained sections were evaluated by a boarded veterinary pathologist. Macroscopic damage in the colons of DSS-treated mice gavaged with P. UF1, ΔlspA P. UF1 or PBS was visualized with a Multi-Purpose Rigid Telescope attached to a TELE PACK X (Karl Storz-Endoscope, Germany), as described previously.13
FITC-dextran gut permeability assay
DSS-treated mice were orally gavaged with FITC-dextran 4000 (Sigma-Aldrich, St. Louis, MO), a nonmetabolizable macromolecule that is used as a permeability probe. All mice were orally gavaged with FITC-dextran (0.6 mg/g mouse weight) and sacrificed 4 h later for serum harvest. Fluorescent intensity in the serum was measured using a microplate reader (BioTek, Winooski, VT) with an excitation wavelength of 485 nm and an emission wavelength of 519 nm. FITC-dextran concentrations in the mouse sera were determined from standard curves generated by serial dilution of FITC-dextran. Serum from mice that were not gavaged with the permeability tracer was used as a control and subtracted from the tested samples.
Statistical analysis
Statistical analyses were performed using GraphPad Prism v7.0. Prior to statistical analysis, normality was tested using the Shapiro-Wilk normality test. Where the groups follow a Gaussian distribution, parametric analyses were performed (2-tailed unpaired t test for 2 variables or one-way ANOVA followed by Tukey’s post-test for 3 variables). Where the groups did not follow a Gaussian distribution, nonparametric analyses were performed (Mann–Whitney U test for 2 variables or Kruskal–Wallis test followed by Dunn’s post-test for 3 variables). P values lower than 0.05 were considered as significant: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
References
Littman, D. R. & Pamer, E. G. Role of the commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe 10, 311–323 (2011).
Lathrop, S. K. et al. Peripheral education of the immune system by colonic commensal microbiota. Nature 478, 250–254 (2011).
Lebeer, S., Vanderleyden, J. & De Keersmaecker, S. C. Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens. Nat. Rev. Microbiol 8, 171–184 (2010).
Akira, S., Uematsu, S. & Takeuchi, O. Pathogen recognition and innate immunity. Cell 124, 783–801 (2006).
Fagan, R. P. & Fairweather, N. F. Biogenesis and functions of bacterial S-layers. Nat. Rev. Microbiol 12, 211–222 (2014).
Thaiss, C. A., Zmora, N., Levy, M. & Elinav, E. The microbiome and innate immunity. Nature 535, 65–74 (2016).
Banchereau, J. & Steinman, R. M. Dendritic cells and the control of immunity. Nature 392, 245–252 (1998).
Medzhitov, R. Recognition of microorganisms and activation of the immune response. Nature 449, 819–826 (2007).
Colliou, N. et al. Commensal Propionibacterium strain UF1 mitigates intestinal inflammation via Th17 cell regulation. J. Clin. Invest. 127, 3970–3986 (2017).
Littman, D. R. & Rudensky, A. Y. Th17 and regulatory T cells in mediating and restraining inflammation. Cell 140, 845–858 (2010).
Colliou, N. et al. Regulation of Th17 cells by P. UF1 against systemic Listeria monocytogenes infection. Gut Microbes. 279–287 (2018).
Ge, Y. et al. Neonatal intestinal immune regulation by the commensal bacterium, P. UF1. Mucosal Immunol. 12, 434–444 (2019).
Lightfoot, Y. L. et al. SIGNR3-dependent immune regulation by Lactobacillus acidophilus surface layer protein A in colitis. EMBO J. 34, 881–895 (2015).
Kordulakova, J. et al. Definition of the first mannosylation step in phosphatidylinositol mannoside synthesis PimA is essential for growth of mycobacteria. J. Biol. Chem. 277, 31335–31344 (2002).
Tatituri, R. V. et al. Inactivation of Corynebacterium glutamicum NCgl0452 and the role of MgtA in the biosynthesis of a novel mannosylated glycolipid involved in lipomannan biosynthesis. J. Biol. Chem. 282, 4561–4572 (2007).
Gentzsch, M. & Tanner, W. The PMT gene family: protein O-glycosylation in Saccharomyces cerevisiae is vital. EMBO J. 15, 5752–5759 (1996).
Herrmann, J. L., OGaora, P., Gallagher, A., Thole, J. E. R. & Young, D. B. Bacterial glycoproteins: A link between glycosylation and proteolytic cleavage of a 19 kDa antigen from Mycobacterium tuberculosis. EMBO J. 15, 3547–3554 (1996).
Sancho, D. & Reis e Sousa, C. Signaling by myeloid C-type lectin receptors in immunity and homeostasis. Annu. Rev. Immunol. 30, 491–529 (2012).
Bekiaris, V., Persson, E. K. & Agace, W. W. Intestinal dendritic cells in the regulation of mucosal immunity. Immunol. Rev. 260, 86–101 (2014).
Schlitzer, A. et al. IRF4 transcription factor-dependent CD11b+ dendritic cells in human and mouse control mucosal IL-17 cytokine responses. Immunity 38, 970–983 (2013).
Price, J. G. et al. CDKN1A regulates Langerhans cell survival and promotes Treg cell generation upon exposure to ionizing irradiation. Nat. Immunol. 16, 1060–1068 (2015).
Cohen, N. et al. GILZ expression in human dendritic cells redirects their maturation and prevents antigen-specific T lymphocyte response. Blood 107, 2037–2044 (2006).
Liu, J. N. et al. The complement inhibitory protein DAF (CD55) suppresses T cell immunity in vivo. J. Exp. Med. 201, 567–577 (2005).
Niess, J. H. & Reinecker, H. C. Lamina propria dendritic cells in the physiology and pathology of the gastrointestinal tract. Curr. Opin. Gastroenterol. 21, 687–691 (2005).
Rescigno, M. & Di Sabatino, A. Dendritic cells in intestinal homeostasis and disease. J. Clin. Invest. 119, 2441–2450 (2009).
Dalod, M., Chelbi, R., Malissen, B. & Lawrence, T. Dendritic cell maturation: functional specialization through signaling specificity and transcriptional programming. EMBO J. 33, 1104–1116 (2014).
Everts, B. et al. Commitment to glycolysis sustains survival of NO-producing inflammatory dendritic cells. Blood 120, 1422–1431 (2012).
Pearce, E. J. & Everts, B. Dendritic cell metabolism. Nat. Rev. Immunol. 15, 18–29 (2015).
Sahu, N. et al. Proline starvation induces unresolved ER stress and hinders mTORC1-dependent tumorigenesis. Cell Metab. 24, 753–761 (2016).
Tang, C. et al. Suppression of IL-17F, but not of IL-17A, provides protection against colitis by inducing Treg cells through modification of the intestinal microbiota. Nat. Immunol. 19, 755–765 (2018).
Lee, J. S. et al. Interleukin-23-independent IL-17 production regulates intestinal epithelial permeability. Immunity 43, 727–738 (2015).
Verma, R. et al. Cell surface polysaccharides of Bifidobacterium bifidum induce the generation of Foxp3(+) regulatory T cells. Sci. Immunol. 3, eaat6975 (2018).
Kamada, N., Seo, S. U., Chen, G. Y. & Nunez, G. Role of the gut microbiota in immunity and inflammatory disease. Nat. Rev. Immunol. 13, 321–335 (2013).
Deutsch, S. M. et al. Identification of proteins involved in the anti-inflammatory properties of Propionibacterium freudenreichii by means of a multi-strain study. Sci. Rep. 7, 46409 (2017).
Le Marechal, C. et al. Surface proteins of Propionibacterium freudenreichii are involved in its anti-inflammatory properties. J. Proteom. 113, 447–461 (2015).
do Carmo, F. L. R. et al. Extractable bacterial surface proteins in probiotic-host interaction. Front. Microbiol. 9, 645 (2018).
do Carmo, F. L. R. et al. Propionibacterium freudenreichii surface protein SlpB is involved in adhesion to intestinal HT-29 cells. Front. Microbiol. 8, 1033 (2017).
Michon, C., Langella, P., Eijsink, V. G., Mathiesen, G. & Chatel, J. M. Display of recombinant proteins at the surface of lactic acid bacteria: strategies and applications. Micro. Cell Fact. 15, 70 (2016).
Ewing, C. P., Andreishcheva, E. & Guerry, P. Functional characterization of flagellin glycosylation in Campylobacter jejuni 81-176. J. Bacteriol. 191, 7086–7093 (2009).
Nothaft, H. & Szymanski, C. M. Protein glycosylation in bacteria: sweeter than ever. Nat. Rev. Microbiol. 8, 765–778 (2010).
Liu, C. F. et al. Bacterial protein-O-mannosylating enzyme is crucial for virulence of Mycobacterium tuberculosis. Proc. Natl Acad. Sci. USA 110, 6560–6565 (2013).
Girrbach, V. & Strahl, S. Members of the evolutionarily conserved PMT family of protein O-mannosyltransferases form distinct protein complexes among themselves. J. Biol. Chem. 278, 12554–12562 (2003).
Geijtenbeek, T. B. & Gringhuis, S. I. C-type lectin receptors in the control of T helper cell differentiation. Nat. Rev. Immunol. 16, 433–448 (2016).
Zhou, Y. et al. Oral tolerance to food-induced systemic anaphylaxis mediated by the C-type lectin SIGNR1. Nat. Med. 16, 1128–1133 (2010).
Lanoue, A. et al. SIGN-R1 contributes to protection against lethal pneumococcal infection in mice. J. Exp. Med. 200, 1383–1393 (2004).
Song, X. et al. Growth factor FGF2 cooperates with interleukin-17 to repair intestinal epithelial damage. Immunity 43, 488–501 (2015).
Kreisman, L. S. & Cobb, B. A. Infection, inflammation and host carbohydrates: a Glyco-Evasion Hypothesis. Glycobiology 22, 1019–1030 (2012).
Dewald, J. H., Colomb, F., Bobowski-Gerard, M., Groux-Degroote, S., Delannoy, P. Role of cytokine-induced glycosylation changes in regulating cell interactions and cell signaling in inflammatory diseases and cancer. Cells 5, 43 (2016).
Gevers, D. et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 15, 382–392 (2014).
Shreiner, A. B., Kao, J. Y. & Young, V. B. The gut microbiome in health and in disease. Curr. Opin. Gastroenterol. 31, 69–75 (2015).
Kaplan, G. G. The global burden of IBD: from 2015 to 2025. Nat. Rev. Gastroenterol. Hepatol. 12, 720–727 (2015).
Li, W. et al. Targeting T cell activation and lupus autoimmune phenotypes by inhibiting glucose transporters. Front Immunol. 10, 833 (2019).
Acknowledgements
This work was supported by NIH R01 DK109560 (to M.M.). Glycomic and glycoproteomic analyses were supported in part by NIH 1S10OD018530 and P41GM10349010 grants (to P.A.) at the Complex Carbohydrate Research Center.
Author information
Authors and Affiliations
Contributions
M.M. directed the experiments, which were executed by Y.G., M.G., M.Z., and J.L. Y.G. and M.Z. performed animal experiments; Y.G. performed flow cytometry analysis, genetic and biochemical assays. Y.G. and J.L. constructed RNA-Seq libraries. M.G. analyzed RNA-Seq data. J.R.A. evaluated and scored all colonic tissue sections. W.L. and L.M. performed extracellular flux analysis. R.S., N.T.S., and P.A. analyzed glycomics and glycoproteomics. M.G., Y.W., D.P.J. and S.L. performed, analyzed, and directed metabolomic studies. Y.G. and M.M. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Ge, Y., Gong, M., Zadeh, M. et al. Regulating colonic dendritic cells by commensal glycosylated large surface layer protein A to sustain gut homeostasis against pathogenic inflammation. Mucosal Immunol 13, 34–46 (2020). https://doi.org/10.1038/s41385-019-0210-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41385-019-0210-0