Summary
Breach of tolerance to gluten leads to the chronic small intestinal enteropathy celiac disease. A key event in celiac disease development is gluten-dependent infiltration of activated cytotoxic intraepithelial lymphocytes (IELs), which cytolyze epithelial cells causing crypt hyperplasia and villous atrophy. The mechanisms leading to gluten-dependent small intestinal IEL infiltration and activation remain elusive. We have demonstrated that under homeostatic conditions in mice, gluten drives the differentiation of anti-inflammatory T cells producing large amounts of the immunosuppressive cytokine interleukin-10 (IL-10). Here we addressed whether this dominant IL-10 axis prevents gluten-dependent infiltration of activated cytotoxic IEL and subsequent small intestinal enteropathy. We demonstrate that IL-10 regulation prevents gluten-induced cytotoxic inflammatory IEL infiltration. In particular, IL-10 suppresses gluten-induced accumulation of a specialized population of cytotoxic CD4+CD8αα+ IEL (CD4+ CTL) expressing Tbx21, Ifng, and Il21, and a disparate non-cytolytic CD4+CD8α− IEL population expressing Il17a, Il21, and Il10. Concomitantly, IL-10 suppresses gluten-dependent small intestinal epithelial hyperproliferation and upregulation of stress-induced molecules on epithelial cells. Remarkably, frequencies of granzyme B+CD4+CD8α+ IEL are increased in pediatric celiac disease patient biopsies. These findings demonstrate that IL-10 is pivotal to prevent gluten-induced small intestinal inflammation and epithelial damage, and imply that CD4+ CTL are potential new players into these processes.
Similar content being viewed by others
Introduction
Celiac disease, a chronic small intestinal enteropathy, is caused by T-cell-driven inflammatory responses to dietary gluten. The only treatment is a lifelong gluten-free diet (GFD), which removes the antigenic trigger and prevents inflammation, but does not reverse the breach of tolerance. Celiac disease results from multiple adaptive and innate immune perturbations culminating in loss of tolerance to gluten. Dysregulation of adaptive immunity is evidenced by the strict major histocompatibility complex (MHC) class II dependency, with patients expressing human leukocyte antigen (HLA)-DQ2 or HLA-DQ8, a genetic susceptibility explained by preferential binding affinity of deamidated gluten peptides to these haplotypes.1 Presentation of deamidated gluten peptides induces differentiation of inflammatory gluten-specific interferon-γ-secreting CD4+ T cells in the lamina propria,2 associated with gluten-specific antibodies (Abs)3 and tissue transglutaminase (TG2) auto-Abs.4 However, T-cell immunity to gluten is not sufficient to cause celiac disease.5,6,7 A second key perturbation is required, namely infiltration of small intestinal cytotoxic intraepithelial lymphocytes (IELs) located at the site of tissue destruction evidenced by crypt hyperplasia and villous atrophy. This IEL infiltration represents a prime feature of celiac disease independent of disease severity and is observed in Marsh 1 lesions where villous blunting is absent.8,9 IEL-infiltrated Marsh 1 lesions also occur in non-celiac gluten sensitivity, a gluten-induced small intestinal inflammation independent of the HLA haplotype and anti-TG2 Abs.10 Moreover, short-term rectal gluten challenge increases mucosal IEL in non-HLA-DQ2/DQ8 siblings of pediatric celiac disease patients.11 However, the mechanisms underlying HLA-independent gluten-specific IEL recruitment are unclear.
IEL, heterogeneous antigen-experienced T cells located between enterocytes, can be subdivided by αβ or γδTCR and CD8αα, CD8αβ, or CD4 expression. Small intestinal IEL are mostly TCRαβ+ CD8αβ+ in humans12 and TCRγδ+ CD8αα+ in mice.13,14 IEL are activated by inflammatory signals and stress-induced molecules to eliminate infected epithelial cells and are thus an essential first-line defense. Massive infiltration of both αβTCR+15 and γδTCR+ IEL are observed in the small intestine in celiac disease.16,17 However, infiltration of αβTCR IEL but not γδTCR IEL correlates with tissue damage severity15,17 and normalizes on a GFD,17 implicating αβTCR IEL in gluten-induced enteropathy.
IEL are activated via ligation of their T-cell receptor (TCR) by cognate antigen18 or in a TCR-independent manner by ligation of activating natural killer receptors (NKRs).19 In celiac disease, TCR-independent IEL activation is driven by two major changes. Interleukin-15 (IL-15) overexpression in inflamed intestinal epithelium5,20 upregulates activating NKR such as NKG2D21 and CD9422 on IEL. Concomitant IFN-γ secretion by gluten-specific CD4+ T cells, leads to overexpression of MHCII23,24 and unconventional MHCI molecules HLA-E and MIC, ligands of CD94/NKG2C and NKG2D, respectively.19,25,26,27,28,29 Ligation of NKR triggers release of cytolytic molecules, granzyme B and perforins, causing epithelial cytolysis and villous atrophy.19,29,30
Despite knowledge of immunological events leading to gluten intolerance, an essential question remains: which mechanism prevents IEL recruitment/activation in gluten-tolerant individuals? Gluten strongly induces T-cell secretion of the anti-inflammatory cytokine IL-10, which may be required to maintain gluten tolerance.7,31,32,33 This prompted us to investigate whether IL-10 signaling controls gluten-dependent IEL infiltration and activation. As IL-10-mediated regulation of T cells requires intact antigen-presenting cell (APC) IL-10 signaling,34,35,36,37 and our previous data showed that mice with deficient IL-10 signaling in APC suffer from spontaneous small intestinal inflammation,34 we assessed the intestinal response to gluten in mice with disrupted IL-10 signaling in CD11c+ APC. Here, we show that disrupting IL-10 signaling in CD11c+ APC leads to a breach of tolerance to gluten and the recruitment of inflammatory cytotoxic CD4+CD8αα+ IEL, associated with a gradual, spontaneous, gluten-dependent small intestinal enteropathy, characterized by epithelial stress—reflected by upregulation of Il15 and unconventional MHCI molecules—and crypt hyperplasia.
Results
Gluten drives IEL infiltration and epithelial stress in the absence of IL-10 signaling
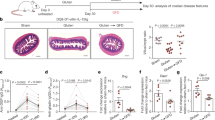
Gluten induces T-cell secretion of the immunomodulatory cytokine IL-10.7,31,32,33 As IL-10 inhibits APC function,34,35,36,37 we assessed whether disrupting IL-10 regulation of CD11c+ cells triggers gluten-dependent IEL infiltration and enteropathy. Hereto, mice lacking the Il10ra gene in CD11c+ cells (ItgaxcreIl10rafl/fl) and co-housed littermates (Il10rafl/fl) were maintained on a gluten-containing (conventional) diet or GFD until sacrifice (30- to 45-week-old). Compared with Il10rafl/fl mice, ItgaxcreIl10rafl/fl mice developed gluten-dependent small intestinal IEL infiltration with increased CD3+ cells lining the duodenal epithelium (Fig. 1a) and increased numbers of CD45+CD3+ IEL (Fig. 1b). Dietary gluten was crucial as both inflammatory features were abolished in ItgaxcreIl10rafl/fl mice on a GFD (Fig. 1a, b). Gluten-induced increase in small intestinal CD45+CD3+ IEL numbers was already observed in 12-week-old ItgaxcreIl10rafl/fl mice (Figure S1A) but the number of CD45+CD3+ IEL was twofold higher in 30- to 45-week-old mice (Fig. 1b and S1A), inferring that gluten-induced IEL accumulation observed in the absence of IL-10 signaling begins at young age and gradually increases with ongoing gluten exposure. Hyperproliferative small intestinal crypts were observed in ItgaxcreIl10rafl/fl mice on a conventional diet, indicated by increased Ki67+ duodenal crypt cells and elevated crypt/villus ratio (Fig. 1c, d). ItgaxcreIl10rafl/fl mice on a GFD had a largely reduced crypt/villus ratio due to crypt hyperplasia, a hallmark of gluten-induced cytolytic IEL activity in celiac disease.13,38 Upregulation of stress-induced molecules and inflammatory cytokines in epithelial cells is a prerequisite for gluten-dependent enteropathy. In the absence of IL-10 signaling, small intestinal CD45neg cells of ItgaxcreIl10rafl/fl mice on a conventional diet had significantly higher expression of MHCII (Fig. 1e, f). In addition, transcription of T23 (encodes stress-induced unconventional MHCI molecule and CD94/NKG2C ligand Qa-1b) was higher in sorted small intestinal EpCAM+ epithelial cells from ItgaxcreIl10rafl/fl mice on a conventional diet (vs. littermates), but not a GFD (Fig. 1g). In contrast, the NKG2D ligand Rae1 was expressed at similar levels in ItgaxcreIl10rafl/fl and littermates irrespective of diet (Fig. 1h). Gluten-dependent epithelial stress and crypt hyperplasia in the absence of IL-10 signaling were T-cell dependent as aged Rag.ItgaxcreIl10rafl/fl mice on a conventional diet did not exhibit crypt hyperplasia or overexpress MHCII on CD45neg cells (vs. littermates; Figure S1B-D). In addition, EpCAM+ cells from ItgaxcreIl10rafl/fl mice on a conventional diet, but not a GFD, had increased levels of Il15 transcripts, a cytokine known to activate IEL in celiac disease19 (Fig. 1i). Of note, serum anti-gluten IgG2c was not elevated in aged ItgaxcreIl10rafl/fl mice on a conventional diet (vs. littermates; Figure S2). These data demonstrate that a single regulatory pathway, namely IL-10 signaling in APC, suppresses gluten-induced small intestinal IEL infiltration, stress-induced NKR ligand upregulation on epithelial cells, and crypt hyperplasia.
Gluten drives CD3+ IEL accumulation, epithelial MHCII expression, unconventional MHCI molecules, and IL-15 upregulation in the absence of IL-10 signaling. ItgaxcreIl10rafl/fl mice and littermates were reared on a gluten-containing (conventional) or gluten-free diet (GFD) until sacrifice. a CD3 immunohistochemistry on duodenal sections. Scale bar, 100 µm; representative of n = 9–21. b Small intestinal CD45+CD3+ IEL number from ItgaxcreIl10rafl/fl mice and littermates obtained by assessing the frequency of single CD45+CD3+ IEL by flow cytometry and multiplying it to the total cell number of the small intestine IEL fraction. c Ki67 immunohistochemistry on duodenal sections. Scale bar, 100 µm; representative of n = 9–21. d Crypt/villus ratio in Ki67-stained sections. e MHCII expression on small intestine single CD45neg cells; representative of n = 6–14. f Geometric mean fluorescence intensity (MFI) of MHCII on single small intestinal CD45neg cells. g–i Quantitative real-time PCR (qRT-PCR) of stress-associated epithelial molecule genes (g, h) or Il15 (i) in sorted live single small intestinal CD45negEpCAM+ cells. Twelve (a), 11 (b-d), 9 (e-f), or 7 independent experiments (g-i). Each square represents one mouse. One-way ANOVA/Bonferroni post-test (d, f: mean + SEM; b, g-i: mean + SD)
Gluten drives inflammatory CD4+ and CD4+CD8α+ IEL infiltration in the absence of IL-10 signaling
Healthy intestinal IEL do not secrete inflammatory cytokines. In celiac disease, infiltrating αβTCR IEL secrete high levels of IFN-γ, which correlates with tissue damage.2,39 In analogy, small intestinal IEL fractions from ItgaxcreIl10rafl/fl mice on a conventional diet, but not a GFD, transcribed more Ifng and Il17a (vs. littermates; Fig. 2a, b). Of note, Il21, a cytokine highly increased during celiac disease,40,41,42 was also higher in IEL of ItgaxcreIl10rafl/fl mice reared on a conventional diet but not on a GFD (Fig. 2c). In keeping with this inflammatory profile, restimulated small intestinal IEL isolated from ItgaxcreIl10rafl/fl mice reared on a conventional diet contained increased frequencies of cells secreting IFN-γ and IL-17 (Fig. 2d, e). At 12 weeks of age, concomitant to IEL infiltration, Ifng and Il17a were increased in ItgaxcreIl10rafl/fl mice on a conventional diet (Figure S3A-B).
Gluten drives inflammatory CD4+ and CD4+CD8α+ IEL accumulation in the absence of IL-10 signaling. ItgaxcreIl10rafl/fl mice and littermates were reared on a conventional or gluten-free diet (GFD) until sacrifice. a-c qRT-PCR of T-cell cytokine genes in total IEL. d Flow cytometry dot plot depicting IL-17 and IFN-γ production by single live CD45+CD3+ IEL isolated from mice reared on conventional diet and restimulated for 4 h with PMA/Ionomycin in the presence of monensin. Representative of three mice per group. e Scatter plot depicting the frequency of IFN-γ and IL-17 producing single live CD45+CD3+ IEL assessed by flow cytometry as in d. f Flow cytometry of CD4 and CD8α on single CD45+CD3+ IEL; representative of n = 8–20. g, h Frequencies (g) and numbers (h) of small intestine IEL (pre-gated on single CD45+CD3+ cells). i CD4 immunohistochemistry on duodenal sections. Scale bar, 100 µm; representative of n = 3–4. Seven (a-c), 2 (d, e), or 13 (f-h) independent experiments. Each square represents one mouse. One-way ANOVA/Bonferroni post-test (mean + SEM) (a-c and g, h) or unpaired Student’s t-test (e) (mean + SD)
Detailed analysis in aged mice revealed that two IEL populations were drastically increased in ItgaxcreIl10rafl/fl mice on a conventional diet (vs. littermates): CD4+CD8α+ IEL increased 28-fold, whereas CD4+CD8α− IEL increased 5.7-fold (Fig. 2f, g). This translated into increased total cell numbers for both these IEL populations (Fig. 2h) and was confirmed by immunohistochemistry showing increased numbers of CD4+ cells lining the epithelium in ItgaxcreIl10rafl/fl mice on a conventional but not a GFD (vs. littermates) (Fig. 2i). The increase in frequency and total cell number was dependent on dietary gluten and normalized on a GFD. The increase in cell numbers but not frequency of CD4+CD8α+ and CD4+CD8α− IEL populations was already observed in 12-week-old ItgaxcreIl10rafl/fl mice on a conventional diet (Figure S3C, D and E). Of note, although the frequency of CD4−CD8α+ IEL was decreased in the absence of IL-10 signaling, the total number of CD4−CD8α+ cells was also increased (Fig. 2g, h).
Functional dichotomy of gluten-dependent CD4+CD8α+ and CD4+ inflammatory IEL
Next, we identified the accumulating IEL population responsible for pro-inflammatory T-cell-derived cytokines. Sorted CD4+CD8α+ IEL from aged ItgaxcreIl10rafl/fl mice on a conventional diet expressed high levels of Ifng but did not express Il17a, whereas CD4+CD8α− IEL expressed significantly less Ifng (Fig. 3a, b) and more Il17a. Interestingly, CD4+CD8α− IEL expressed more Il10 than CD4+CD8α+ IEL, whose Il10 expression was very low (Fig. 3d), indicating CD4+CD8α+ IEL have a more pro-inflammatory phenotype. Such a division of labor was not seen for expression of the celiac disease-associated pro-inflammatory cytokine Il21 as both CD4+CD8α− and CD4+CD8α+ IEL expressed high levels of Il21 (Fig. 3c). Notably, all four cytokines were expressed at lower levels in CD4−CD8α+ classical cytotoxic IEL than either CD4+ population (Fig. 3a-d). In accordance with their high Ifng and very low Il17a expression, CD4+CD8α+ IEL expressed higher levels of Tbx21 (encoding for T-bet) than CD4+CD8α− IEL (Fig. 3e), which correlated with T-bet positivity and RORγt negativity measured by flow cytometry (Fig. 3f, g). Similarly, the population of CD4+CD8α− IEL expressing high levels of Il17a and lower levels of Ifng was mostly RORγt+ and partly T-bet+ (Fig. 3f, g). Approximately 20% of CD4+CD8α− IEL (Il10 high) expressed Foxp3, whereas most CD4+CD8α+ IEL (Il10 low) were Foxp3− (Fig. 3h). Therefore, gluten leads to accumulation of two phenotypically and functionally distinct IEL populations in the absence of IL-10 signaling: CD4+CD8α+ IEL mainly expressing Ifng and Il21, and CD4+CD8α− IEL with a mixed phenotype of regulatory Il10 and inflammatory Il17a and Il21 expressing cells.
Functionally distinct CD4+CD8α+ and CD4+ IEL accumulate in the absence of IL-10 signaling. ItgaxcreIl10rafl/fl mice were reared on a conventional diet until sacrifice. a-e qRT-PCR of cytokine genes and associated transcription factors on sorted live single small intestine IEL (n = 4). f-g Flow cytometry of T-bet (f) and RORγt (g) on small intestine IEL (pre-gated on single CD45+CD3+); representative of n = 6. h Frequencies of Foxp3+ cells in small intestine IEL (pre-gated on single CD45+CD3+). Each square represents one mouse. Three (a-e, h) or two independent experiments (f-g). One-way ANOVA/Bonferroni post-test (mean + SD)
CD4+CD8α+ IEL accumulating after continuous gluten ingestion in the absence of IL-10 signaling are cytotoxic CD4+ CTL
Small intestinal CD4+CD8α+ IEL derive from CD4+ αβTCR+ MHCII-restricted T helper cells.43,44 Repetitive exposure to cognate antigens induces transcriptional reprogramming of CD4+ T helper cells to CD4+ cytotoxic T lymphocytes (CD4+ CTL), involving ThPOK downregulation, Runx3 upregulation and initiation of a cytolytic program.43,45 In the presence of IL-15, CD4+ CTL produce IFN-γ and become highly cytolytic to MHCII-expressing cells.43 Indicative of transcriptional reprogramming, sorted CD4+CD8α+ IEL accumulating in response to gluten in ItgaxcreIl10rafl/fl mice expressed lower levels of Thpok (encoded by Zbtb7b) and higher levels of Runx3 compared with CD4+CD8α− IEL both at transcript and protein level (Fig. 4a-d). In support of this idea, gluten-dependent CD4+CD8α+ IEL did not express CD8β, hence are CD8αα homodimer positive (Fig. 4e), a characteristic feature of CD4+ CTL, and carry CD4+ CTL-associated marker CD103 (Fig. 4f) and the cytotoxicity-associated molecule 2B4 (CD244) (Fig. 4g). CD4+CD8α+ IEL of ItgaxcreIl10rafl/fl mice on a conventional diet transcribed significantly more Crtam than CD4+CD8α− IEL, a molecule that plays an essential role in classical CD4+ T helper cell differentiation to CD4+ CTL46,47 (Fig. 4h). Moreover, in contrast to CD4−CD8α+ classical IEL, both small intestinal CD4+CD8α+ and CD4+CD8α− IEL of ItgaxcreIl10rafl/fl mice on a conventional diet lacked γδTCR (Fig. 4i) strongly suggesting CD4+CD8α+ IEL derive from CD4+ T helper cells. These data establish that, in the absence of IL-10 signaling, gluten drives intraepithelial infiltration of CD4+CD8α− IEL, as well as CD4+CD8α+ IEL with phenotypic characteristics of CD4+ CTL. This, in combination with increased expression of both MHCII and Qa-1b (Fig. 1e-g) led us to hypothesize that CD4+CD8α+ IEL accumulating in Itgaxcre Il10rafl/fl mice on a gluten-containing diet could be involved in cytolysis of epithelial cells underpinning crypt hyperplasia. CD4+CD8α+ IEL from ItgaxcreIl10rafl/fl mice on a conventional diet expressed more Gzmb and Prf1 (encoding Granzyme B and perforin, respectively) than CD4+CD8α− or classical CD4−CD8α+ IEL (Fig. 5a, b). Granzyme B protein frequency and mean fluorescence intensity (MFI) were higher in CD4+CD8α+ IEL than CD4+CD8α− IEL and Granzyme B frequency was equal in CD4+CD8α+ IEL and CD4−CD8α+ classical cytotoxic IEL (Fig. 5c-e). Neither the frequency nor MFI of NKG2D were increased on CD4+CD8α+ IEL compared with CD4+CD8α− IEL (Fig. 5f, g), in line with a lack of Rae1 expression (NKG2D ligand) on epithelial cells (Fig. 1h), suggesting NKG2D ligation does not participate in IEL activation leading to crypt hyperplasia in ItgaxcreIl10rafl/fl mice. In agreement with high Granzyme B and perforin expression, CD4+CD8α+ IEL from ItgaxcreIl10rafl/fl mice on a conventional diet had a higher frequency of CD107a expression, a surrogate marker for cytolysis, and higher CD107a MFI than CD4+CD8α− IEL after TCRβ restimulation (Fig. 5h-j; Supplementary Table 3), unequivocally demonstrating cytotoxic activity of gluten-dependent CD4+ CTL accumulating in the absence of IL-10 signaling. Altogether, this demonstrates that IL-10 signaling in APC suppresses gluten-induced infiltration of CD4+ CTL.
CD4+CD8α+ IEL accumulating in the absence of IL-10 signaling after continuous gluten ingestion have a CD4+ CTL phenotype. ItgaxcreIl10rafl/fl mice were reared on a conventional diet. a, b qRT-PCR of Zbtb7b (a) and Runx3 (b) on sorted small intestine live single CD45+CD3+CD4+CD8α+, CD45+CD3+CD4+CD8α−, CD45+CD3+CD4−CD8α+ IEL (n = 4). c-g Flow cytometry of Thpok (c), Runx3 (d), CD8α and CD8β (e), CD103 (f) and 2B4 (g) on small intestine IEL (pre-gated on single CD45+CD3+); representative of n = 3 (c), 6 (d and g), 8 (f) or 13 (e). h qRT-PCR for Crtam on sorted small intestine live single CD45+CD3+CD4+CD8α+, CD45+CD3+CD4+CD8α−, CD45+CD3+CD4−CD8α+ IEL (n = 4). i Flow cytometry of γδTCR on small intestine IEL (pre-gated on single CD45+CD3+); representative of n = 8. One (c), two (d, g), four (a-b, h), or five independent experiments (e-f, i). One-way ANOVA/Bonferroni post-test (mean + SEM)
Cytotoxic CD4+CD8α+ IEL accumulate in the absence of IL-10 signaling after continuous gluten ingestion. ItgaxcreIl10rafl/fl mice were reared on a conventional diet. a, b qRT-PCR for genes encoding cytotoxic molecules on sorted live small intestine single CD45+CD3+CD4+CD8α+, CD45+CD3+CD4+CD8α−, CD45+CD3+CD4−CD8α+ IEL (n = 4). c Histogram depicting Granzyme B staining on CD45+CD3+CD4+CD8α+, CD45+CD3+CD4+CD8α−, CD45+CD3+CD4−CD8α+ IEL of a ItgaxcreIl10rafl/fl mouse reared on a gluten-containing diet (representative of n = 8). d-g Frequency (d, f) and geometric mean fluorescence intensity (MFI; e, g) of Granzyme B and NKG2D assessed by flow cytometry (pre-gated on single CD45+CD3+). h Flow cytometric analysis of CD107a IEL isolated from mice reared on a conventional diet and stimulated either with an isotype or with an anti-βTCR Ab. (i-j) Frequency (i) and MFI (j) of CD107a of small intestine IEL assessed by flow cytometry (pre-gated on single CD45+CD3+). Frequency and MFI obtained by subtracting the frequency (or MFI) of unstimulated IEL from that of αβTCR-stimulated IEL. Each square represents one mouse. Three (a-b), five (c-g), or two independent experiments (h-j). One-way ANOVA/Bonferroni post-test (a-j) or unpaired Student’s t-test (h); a, b and i: mean + SD; d-f: mean + SEM
Gluten selectively drives accumulation of CD4+CD8α+ LPLs in the absence of IL-10 regulation
As CD4+ CTL arise from classical CD4+ T helper cells under repetitive presentation of cognate antigens by APC,43 gluten-dependent infiltration of CD4+ CTL in ItgaxcreIl10rafl/fl mice should be associated with increased CD4+CD8α+ T-cell infiltration in the lamina propria. The CD4+CD8α+ frequency in small intestinal lamina propria lymphocytes (LPLs) was significantly higher in aged ItgaxcreIl10rafl/fl mice on a conventional diet (vs. littermates, Fig. 6a, b) but—similarly to IEL frequency—not at 12 weeks of age (Figure S4A-B). The CD4+CD8α− LPL frequency was also increased in aged ItgaxcreIl10rafl/fl mice on both a conventional and a GFD (Fig. 6a, b) showing that gluten alone drives CD4+CD8α+ LPL accumulation when IL-10 control is lost, but that CD4+CD8α− LPL accumulation is dependent on other antigenic signals. Similarly to the IEL fraction, both Ifng and Il17a were upregulated in the total LPL fraction of 12-week-old (Figure S4C-D) and aged ItgaxcreIl10rafl/fl mice on a conventional diet compared with littermates (Fig. 6c, d). Cytokine expression by LPL in ItgaxcreIl10rafl/fl mice on a conventional diet mirrored the IEL populations. CD4+CD8α+ LPL expressed more Ifng but less Il17a and Il10 than CD4+CD8α− LPL (Fig. 6e-g). CD4+CD8α− LPL (which accumulate even on a GFD) expressed the highest levels of Il17a, reflecting that upregulation of Il17a in the total LPL fraction was not abolished by a GFD (Fig. 6d). Similarly, high Ifng expression by CD4+CD8α+, whose accumulation in the absence of IL-10 signaling was prevented by a GFD, correlated with absence of Ifng increase in the total LPL fraction of ItgaxcreIl10rafl/fl mice on a GFD (Fig. 6c). Of note, absence of IL-10 signaling did not affect the Foxp3+ CD4+ LPL frequency (Figure S5A). CD4+CD8α+ LPL also displayed evidence of transcriptional reprogramming into CD4+ CTL; they expressed the homodimer CD8αα and integrin CD103, tended to transcribe less Zbtb7b and more Runx3 and Crtam than CD4+CD8α− LPL, and expressed 2B4 while maintaining surface αβTCR (Figure S5B-H). Moreover, CD4+CD8α+ LPL expressed more Prf1 and Gzmb and had a significantly higher Granzyme B+ frequency and MFI than CD4+CD8α− LPL (Figure S6A-D), demonstrating that transcriptional modifications are associated with cytolytic properties. Therefore, in the absence of IL-10 signaling gluten drives accumulation of CD4+CD8α+ T cells with CD4+ CTL characteristics in the lamina propria.
CD4+CD8α− and CD4+CD8α+ T cells accumulate in the small intestinal lamina propria in the absence of IL-10 signaling, but only CD4+CD8α+ accumulation solely depends on gluten. ItgaxcreIl10rafl/fl mice and littermates were reared on a conventional or GFD until sacrifice. a Flow cytometry of CD4 and CD8α on small intestine lamina propria lymphocytes (LPLs) (pre-gated on single CD45+CD3+); representative of n = 7–20. b Frequencies of single small intestine LPL assessed by flow cytometry (pre-gated on single CD45+CD3+). c-g qRT-PCR (n = 3) of T-cell-derived cytokine genes in total LPL fraction (c, d) and sorted live single small intestine CD45+CD3+CD4+CD8α+, CD45+CD3+CD4+CD8α−, CD45+CD3+CD4−CD8α+ LPL (e-g). Each square represents one mouse. Twelve (a, b), four (c, d), or three independent experiments (e-g). One-way ANOVA/Bonferroni post-test (mean + SD)
Granzyme B+ CD4+CD8αα+ IEL are present in pediatric celiac disease
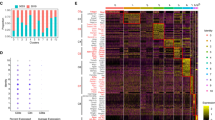
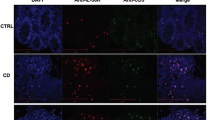
To determine whether CD4+ CTL could be involved in gluten-dependent small intestinal inflammation and damage in human, we determined whether granzyme B+ CD4+CD8αα+ IEL could be detected in pediatric celiac disease. Biopsies from celiac disease patients with Marsh 3 lesions were compared with biopsies of patients undergoing endoscopy due to suspicion of celiac disease but for whom IEL infiltration and normal mucosal morphology was found (Marsh 0). CD4+CD8α− and CD4+CD8αα+ IEL expressing αβTCR were detected in all biopsies (Fig. 7a). However, the CD4+CD8α+ granzyme B+ frequency was significantly higher in Marsh 3 patients than Marsh 0 patients (Fig. 7b, c) with four out of the nine Marsh 3 patients displaying higher CD4+CD8α+ granzyme B+ frequency when compared with the seven Marsh 0 patients. CD4+CD8+ IEL were also detectable by immunofluorescence in Marsh 3 celiac disease lesions (Figure S7A-B). Together, these data strongly argue that gluten-dependent CD4+ CTL may participate in celiac disease.
Granzyme B+ CD4+CD8α+ IEL are detectable in human pediatric celiac disease duodenal tissue. a Flow cytometry of CD8β and αβTCR on CD4+CD8α+ IEL (pre-gated on live CD45+CD3+) in duodenal biopsies from a Marsh 3 pediatric celiac disease patient; representative of n = 9. b Dot plot depicting Granzyme B production by CD4−CD8α+ and CD4+CD8α+ IEL of a Marsh 0 and a Marsh 3 celiac disease patient; representative of n = 7 and 9, respectively. c. Frequency of Granzyme B+ CD4+CD8α+, Granzyme B+ CD4+CD8α− IEL and Granzyme B+ CD4−CD8α+ IEL in Marsh 0 and Marsh 3 patients assessed by flow cytometry. Each square represents one patient. Eight independent experiments. Mann–Whitney U-test
Discussion
In celiac disease, aberrant responses to gluten culminate in cytotoxic IEL infiltration and killing of stressed small intestine epithelial cells, causing crypt hyperplasia and villous atrophy. Despite knowledge of the faulty HLA-DQ2/8-dependent gluten-specific inflammatory CD4+ T-cell response, the mechanisms underlying gluten-induced cytotoxic IEL infiltration and subsequent tissue destruction remain elusive. We demonstrate that in mice, constitutive IL-10 regulation of APC prevents gluten-induced cytotoxic inflammatory IEL infiltration, epithelial activation (evidenced by upregulation of Il15 and T23), and crypt hyperplasia. In particular, IL-10 suppresses gluten-induced accumulation of cytotoxic CD4+CD8α+ IEL (CD4+ CTL) expressing Tbx21 and Ifng, as well as disparate non-cytolytic CD4+CD8α− IEL expressing Il17a and Il10. Gluten-dependent enteropathy develops gradually with age independently of the HLA-DQ2/8 haplotype and is not associated with anti-TG2 Abs, a hallmark autoimmune feature that depends on HLA-DQ2/8-restricted gluten-specific conventional CD4+ T cells (Figure S7C). Our findings reinforce the notion that both HLA-dependent and HLA-independent immune defects are concomitantly required for full-blown celiac disease. Disrupting IL-10 signaling triggered gluten-induced epithelial stress with upregulation of IL-15, MHCII, and Qa-1b, an activating ligand for the NKR CD94,22 which altogether license IEL to perform cytolysis and is likely due to increased production of IFN-γ that upregulates stress molecules on epithelial cells.23,24 Importantly, Rae1 (NKG2D ligand) and NKG2D were not upregulated on CD4+ CTL in the absence of IL-10 signaling. This additional epithelial stress may be required to induce maximal CD4+ and CD8+ CTL cytolytic capacity and villous atrophy.
Gluten is highly degradation resistant, which allows translocation of large immunogenic peptides to the lamina propria.48 Despite its immunogenic properties, gluten is tolerated in most individuals; this may partly rely on the capacity of gluten to induce T-cell IL-10 production as reported in mouse models and human celiac disease.7,31,32 Here we demonstrate that IL-10 regulation of APC is pivotal to control gluten-induced IEL infiltration and crypt hyperplasia. IL-10 primarily exerts its anti-inflammatory role via APC by preventing pro-inflammatory cytokine release,34,36,37 suggesting that the effects of gluten on APC could be drastically exacerbated in the absence of IL-10 regulation. In turn, APC activation could lead to IFN-γ producing CD4+ T-cell differentiation and conversion to CD4+ CTL. Alternatively, gluten could indirectly alter small intestinal microbiota composition, which would in turn induce inflammatory responses in the absence of IL-10 regulation. Preliminary microbiota composition analysis of the feces of ItgaxcreIl10rafl/fl mice and littermates did not reveal obvious differences (data not shown); further analyses are required to determine whether gluten directly affects APC or induces microbiota alterations in the absence of IL-10 signaling. A recent study demonstrated that Lactobacillus reuteri induces CD4+CD8α+ IEL by tryptophan-mediated aryl-hydrocarbon receptor activation and subsequent conversion of CD4+ T cells into CD4+CD8α+ IEL.49 In our colony, L. reuteri was detected in the feces of both Il10rafl/fl mice and ItgaxcreIl10rafl/fl mice irrespective of diet (Figure S8). Therefore, we do not anticipate that the large accumulation of CD4+CD8αα+ IEL in the absence of IL-10 regulation of APCs is due to selective colonization of L. reuteri in ItgaxcreIl10rafl/fl mice. Of note, although IEL infiltration was completely prevented in ItgaxcreIl10rafl/fl mice on a GFD, these animals still had signs of low-grade inflammation (mild crypt hyperplasia and increased frequency of Ifng-expressing CD4+ T helper cells) in lamina propria possibly due to other food or microbial triggers.
We provide the first evidence of gluten-driven CD4+ CTL conversion, and the first identification of granzyme B+ CD4+CD8αα+ IEL in pediatric human celiac disease biopsies. The MHCII-restricted cytolytic capacity of CD4+ CTL, initially identified during viral infections, is beneficial for host defense.50 However, increased CD4+ CTL frequencies are observed in the synovia in rheumatoid arthritis51 and circulation in ankylosing spondylitis52 and associate with disease severity, suggesting a pathogenic role of these cytolytic cells during chronic inflammation. Together, these data argue for a role of CD4+ CTL either in the onset or maintenance of gluten-dependent chronic inflammation. Repeated antigen recognition induces CD4+ CTL conversion and coincides with ThPOK loss and CD8α, Runx3 and granzyme B upregulation.43 This agrees with the gradual development of gluten-dependent CD4+ CTL accumulation in our model and is in line with a role for these cells in chronic inflammatory diseases. Antigen presence alone is not sufficient to induce cytolytic activity of tissue-resident CD4+ CTL.43 Antigen reactivation in the presence of IL-15 enhances CD4+ CTL activation.43 As deficient IL-10 regulation in our model was associated with gluten-dependent epithelial IL-15 upregulation, further investigation should reveal whether IL-15 is involved in gluten-driven CD4+ CTL activation. A role for TGF-β was recently revealed in the differentiation of intestinal CD4+ CTL by CD103+CD11b− dendritic cells (DCs).46,53 As we have previously demonstrated that CD103+CD11b− DC preferentially take up dietary gluten in the small intestine when compared with other DC subpopulations and macrophages,32,54 this raises the question whether IL-10 regulation of CD103+CD11b− DC critically prevents gluten-induced CD4+ CTL infiltration in ItgaxcreIl10rafl/fl mice. However, as the cre/lox system we used here targets all CD11c-expressing APC and since we have shown that macrophages play an important role in degrading gluten,32 unraveling the relative contribution of individual intestinal APC populations to the gluten-induced conversion of CD4+ T helper cells into CD4+ CTL will require further analysis.
Altogether our study demonstrates that one single biological pathway, IL-10 signaling in APC, prevents gluten-dependent infiltration of CD4+ CTL, as well as epithelial activation. Deregulation of IL-10-mediated control of APC likely represents a previously unappreciated early step in the cascade of events culminating in celiac disease.
Experimental procedures
Mice
ItgaxcreIl10rafl/fl mice, Rag.ItgaxcreIl10rafl/fl mice with deletion of the Il10ra in CD11c+ cells, and littermate controls (Il10rafl/fl mice and Rag.Il10rafl/fl mice, respectively) were described previously.34 In brief, Itgaxcre mice were crossed to Il10rafl/fl mice to obtain ItgaxcreIl10rafl/fl. ItgaxcreIl10rafl/fl mice were crossed to Rag2-deficient mice (Jackson Laboratories) to generate Rag.ItgaxcreIl10rafl/fl double knockout mice. All mice were on a C57/Bl6 background. All mice were reared under specific pathogen-free conditions in individually ventilated cages. Water and food were provided ad libitum. All experiments were approved by and performed in accordance with the animal experimental committee of Erasmus Medical Center, Rotterdam, The Netherlands. Upon weaning and until sacrifice, mice were either fed a conventional chow diet (SDSdiets) or GFD (ABdiets). ItgaxcreIl10rafl/fl mice and l10rafl/fl mice from the same litter were randomly allocated to the diets. Unless otherwise indicated, mice were sacrificed at 30–45 weeks. Both males and females were used in this study. As previously reported,53 a germ-line deletion was observed yielding Il10rafl/ko mice. No differences were observed between Il10rafl/fl mice and Il10rafl/ko mice in any experiments (data not shown); therefore, Il10rafl/fl mice and Il10rafl/ko mice were pooled for analysis and all denominated Il10rafl/fl throughout the article.
Patients
Duodenal biopsies were obtained from pediatric patients (aged 2–16 years) during diagnostic upper endoscopy performed at Maasstad Hospital or Erasmus University Medical Centre, Rotterdam, The Netherlands. Studies were approved by the medical ethical committee of the Erasmus Medical Centre and the parents of all participants gave written informed consent before enrollment. No patients were following a GFD prior to endoscopy. Celiac disease diagnoses and Marsh scores were determined by a pathologist during routine diagnostic work-up.
Murine intraepithelial and LPL isolation
Small intestinal IELs and LPLs were isolated as previously described.34 Whole small intestinal tissue was excised and incubated twice in Hank’s balanced salt solution (HBSS) (Gibco) supplemented with 10% fetal calf serum (FCS; Bodinco), 5 mM EDTA (Sigma), and 1 mM DTT (Sigma) for 20 min at 37 °C with agitation. The supernatant containing IEL was washed and resuspended in 44% percoll (Pharmacia), overlaid with 67% percoll, and centrifuged for 20 min at 684 g. IEL were recovered from the interface layer. The remaining intestinal segments, containing LP cells, were digested twice in RPMI supplemented with 10% FCS, 100 U/mL collagenase type VIII (Sigma) and 0.01 mg/mL DNAse (Sigma) for 1 h at 37 °C with agitation. Cells were passed through a 70 μm nylon cell strainer, resuspended in 100% percoll, overlayered with 40% percoll, centrifuged for 20 min at 684 g and LPL were recovered from the interface layer.
Human IEL isolation
Freshly obtained biopsies were incubated twice in RPMI supplemented with 1% FCS, 250 mM EGTA, and 45 mM MgCl2 for 30 min at 37 °C with agitation. Cells were passed through a 40 µm cell strainer and washed with RPMI.
RNA isolation and cDNA synthesis
Total RNA was extracted using the Nucleospin RNA-XS or RNAII kit (Macherey-Nagel) and reverse transcribed to complementary DNA (cDNA) using random hexamers (2.5 µM), oligo(dT) primers (20 nM), dNTP (0.2 mM), M-MLV (200 units, Promega), and RNAsin (25 units, Promega).
Quantitative PCR
Real-time quantitative polymerase chain reaction (qRT-PCR) was performed using general fluorescence-based detection with SYBR green on an AbiPrismR 7900 Sequence Detection System (PE Applied Biosystems). Cyclophillin was used as a reference gene to control for sample loading and enable normalization between samples. Target gene expression levels relative to Cyclophilin were calculated using: relative expression level = 2−ΔCt, whereby ΔCt = Cttarget–Ctcyclophilin. Specific primers spanning different constant region exons were designed (Table S1).
Flow cytometry, fluorescence-activated cell sorting, and Abs
Single-cell suspensions were incubated with normal mouse serum for 10 min on ice followed by a 30-min incubation with the Ab mix on ice. For analysis of intracellular factors, cells were fixed and permeabilized using the Foxp3 permeabilisation kit (ebioscience). Abs used are described in Table S2. Dead cells were excluded using 7AAD (Invitrogen), 4',6-diamidino-2-phenylindole (DAPI) (Molecular Probes), or a fixable Live/dead dye (ThermoFisher). Flow cytometry data were acquired using a FACS Canto II (BD) and analyzed using FlowJo software (LLC). A FACSAria (BD) was used for fluorescence-activated cell sorting.
Immunohistochemistry
Murine small intestinal tissues were fixed in 4% paraformaldehyde (PFA), paraffin embedded and 4 µm thick sections were stained with hematoxylin (Vector Laboratories) and eosin (Sigma). For detection of mCD3 (polyclonal, Dako) and mKi67 (TEC-3, Dako), endogenous peroxidase activity was quenched with 1.5% H2O2 in phosphate buffered saline (PBS) for 30 min, followed by microwave treatment in citrate buffer (10 mM, pH 6.0) for antigen retrieval. For detection of mCD4 (4SM95, ebioscience), antigen retrieval was achieved by pepsin pretreatment (0.1% in 0.01 N HCl at 37 °C). Sections were blocked for 1 h in 10 mM Tris, 5 mM EDTA, 0.15 M NaCl, 0.05% Tween-20, 0.25% gelatin, and 10% normal mouse serum (for mCD3 and mKi67) or 10% normal mouse serum and 10% normal donkey serum (for mCD4), then incubated overnight with primary Ab at 4 °C. Immunoreactivity were detected using biotinylated secondary goat anti-rabbit Ab (for CD3 and Ki67) or donkey anti-rat Ab (for CD4) using the Vectastain ABC Elite Kit (Vector Laboratories) and 3,3′-diaminobenzidine tetrahydrochloride (Sigma-Aldrich). Sections were counterstained with hematoxylin. For detection of hCD4 (4B12, ThermoFisher Scientific) and hCD8 (SP12, ThermoFisher Scientific), human duodenal biopsies were fixed in 4% PFA, paraffin embedded, 4 µm thick sections were microwaved in citrate buffer (10 mM, pH 6.0) for antigen retrieval, blocked for 1 h in 10 mM Tris, 5 mM EDTA, 0.15 M NaCl, 0.05% Tween-20, 0.25% gelatin, 10% normal horse serum, and 10% normal goat serum in a 20% solution #1 of Streptavidin/Biotin Blocking Kit (Vector Laboratories), incubated overnight at 4 °C in primary Ab diluted in PBS containing 2% normal human serum in a 20% solution #2 of Streptavidin/Biotin Blocking Kit (Vector Laboratories). The next day, slides were incubated for 1 h with biotinylated horse anti-mouse IgG (Vector Laboratories) in PBS with 2% goat serum, incubated for 1 h with DyLight 594 Streptavidin (Vector Laboratories) and DyLight 488 Goat anti-rabbit IgG (Vector Laboratories) in PBS. Slides were stained with DAPI (Sigma-Aldrich), mounted in Mowiol (Sigma), and images were acquired using a Leica DM5500B upright microscope and LAS-AF image acquisition software (Leica Microsystems).
CD107 mobilization assay
IEL were isolated as described above and enriched for CD45+ cells by negative selection using an anti-EpCAM Ab and Dynabeads technology (Life Technologies) according to the manufacturer’s recommendations. In brief, the total IEL fraction was incubated with purified rat IgG anti-mouse EpCAM Ab (G8.8, Biolegend), followed by incubation with sheep anti-rat IgG Dynabeads (Life Technologies). Bead-bound EpCAM-labeled cells were removed using a Dynabead magnet. The EpCAM-negative fraction (enriched in CD45+ cells) was collected and plated in a 96-well U-bottom plate (previously coated with anti-TCRβ Ab (2 µg/mL; Biolegend)) in the presence of Golgi plug (BD) and CD107a Ab (ebioscience) for 4 h at 37 °C and 5% CO2.
Statistical analysis
GraphPad Prism version 5 was used to create graphs and perform statistical analysis. Normally distributed data sets (assessed by a Kolmogorov–Smirnov test) were analyzed using a two-tailed unpaired or paired Student’s t-test (comparison of two groups) or one-way analysis of variance (ANOVA; three or more groups) followed by the Bonferroni post-test. Non-normally distributed data sets were analyzed using the Mann–Whitney U-test (comparison of two groups); *p < 0.05; **p < 0.01; ***p < 0.001.
References
Lundin, K. E. et al. Gliadin-specific, HLA-DQ(alpha 1*0501,beta 1*0201) restricted T cells isolated from the small intestinal mucosa of celiac disease patients. J. Exp. Med. 178, 187–196 (1993).
Nilsen, E. M. et al. Gluten specific, HLA-DQ restricted T cells from coeliac mucosa produce cytokines with Th1 or Th0 profile dominated by interferon gamma. Gut 37, 766–776 (1995).
Volta, U. et al. Antibodies to gliadin detected by immunofluorescence and a micro-ELISA method: markers of active childhood and adult coeliac disease. Gut 26, 667–671 (1985).
Dieterich, W. et al. Identification of tissue transglutaminase as the autoantigen of celiac disease. Nat. Med. 3, 797–801 (1997).
Setty, M. et al. Distinct and synergistic contributions of epithelial stress and adaptive immunity to functions of intraepithelial killer cells and active celiac disease. Gastroenterology 149, 681–691e10 (2015).
de Kauwe, A. L. et al. Resistance to celiac disease in humanized HLA-DR3-DQ2-transgenic mice expressing specific anti-gliadin CD4 + T cells. J. Immunol. 182, 7440–7450 (2009).
Du Pre, M. F. et al. Tolerance to ingested deamidated gliadin in mice is maintained by splenic, type 1 regulatory T cells. Gastroenterology 141, 610–620 (2011). 20 e1-2.
Husby, S. et al. European Society for Pediatric Gastroenterology, Hepatology, and Nutrition guidelines for the diagnosis of coeliac disease. J. Pediatr. Gastroenterol. Nutr. 54, 136–160 (2012).
Marsh, M. N. Grains of truth: evolutionary changes in small intestinal mucosa in response to environmental antigen challenge. Gut 31, 111–114 (1990).
Catassi, C. et al. Diagnosis of non-celiac gluten sensitivity (NCGS): the Salerno Experts’ Criteria. Nutrients 7, 4966–4977 (2015).
Troncone, R. et al. In siblings of celiac children, rectal gluten challenge reveals gluten sensitization not restricted to celiac HLA. Gastroenterology 111, 318–324 (1996).
Jarry, A., Cerf-Bensussan, N., Brousse, N., Selz, F. & Guy-Grand, D. Subsets of CD3 + (T cell receptor alpha/beta or gamma/delta) and CD3- lymphocytes isolated from normal human gut epithelium display phenotypical features different from their counterparts in peripheral blood. Eur. J. Immunol. 20, 1097–1103 (1990).
Abadie, V., Discepolo, V. & Jabri, B. Intraepithelial lymphocytes in celiac disease immunopathology. Semin. Immunopathol. 34, 551–566 (2012).
Cheroutre, H., Lambolez, F. & Mucida, D. The light and dark sides of intestinal intraepithelial lymphocytes. Nat. Rev. Immunol. 11, 445–456 (2011).
Kutlu, T. et al. Numbers of T cell receptor (TCR) alpha beta + but not of TcR gamma delta + intraepithelial lymphocytes correlate with the grade of villous atrophy in coeliac patients on a long term normal diet. Gut 34, 208–214 (1993).
Halstensen, T. S., Scott, H. & Brandtzaeg, P. Intraepithelial T cells of the TcR gamma/delta + CD8- and V delta 1/J delta 1 + phenotypes are increased in coeliac disease. Scand. J. Immunol. 30, 665–672 (1989).
Calleja, S. et al. Dynamics of non-conventional intraepithelial lymphocytes-NK, NKT, and gammadelta T-in celiac disease: relationship with age, diet, and histopathology. Dig. Dis. Sci. 56, 2042–2049 (2011).
Guy-Grand, D., Cuenod-Jabri, B., Malassis-Seris, M., Selz, F. & Vassalli, P. Complexity of the mouse gut T cell immune system: identification of two distinct natural killer T cell intraepithelial lineages. Eur. J. Immunol. 26, 2248–2256 (1996).
Meresse, B. et al. Coordinated induction by IL15 of a TCR-independent NKG2D signaling pathway converts CTL into lymphokine-activated killer cells in celiac disease. Immunity 21, 357–366 (2004).
Mention, J. J. et al. Interleukin 15: a key to disrupted intraepithelial lymphocyte homeostasis and lymphomagenesis in celiac disease. Gastroenterology 125, 730–745 (2003).
Roberts, A. I. et al. NKG2D receptors induced by IL-15 costimulate CD28-negative effector CTL in the tissue microenvironment. J. Immunol. 167, 5527–5530 (2001).
Jabri, B. et al. Selective expansion of intraepithelial lymphocytes expressing the HLA-E-specific natural killer receptor CD94 in celiac disease. Gastroenterology 118, 867–879 (2000).
Kvale, D., Brandtzaeg, P. & Lovhaug, D. Up-regulation of the expression of secretory component and HLA molecules in a human colonic cell line by tumour necrosis factor-alpha and gamma interferon. Scand. J. Immunol. 28, 351–357 (1988).
Scott, H., Sollid, L. M., Fausa, O., Brandtzaeg, P. & Thorsby, E. Expression of major histocompatibility complex class II subregion products by jejunal epithelium in patients with coeliac disease. Scand. J. Immunol. 26, 563–571 (1987).
Braud, V. M. et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 391, 795–799 (1998).
Bauer, S. et al. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science 285, 727–729 (1999).
Groh, V. et al. Cell stress-regulated human major histocompatibility complex class I gene expressed in gastrointestinal epithelium. Proc. Natl. Acad. Sci. USA 93, 12445–12450 (1996).
Groh, V., Steinle, A., Bauer, S. & Spies, T. Recognition of stress-induced MHC molecules by intestinal epithelial gammadelta T cells. Science 279, 1737–1740 (1998).
Hue, S. et al. A direct role for NKG2D/MICA interaction in villous atrophy during celiac disease. Immunity 21, 367–377 (2004).
Di Sabatino, A. et al. Epithelium derived interleukin 15 regulates intraepithelial lymphocyte Th1 cytokine production, cytotoxicity, and survival in coeliac disease. Gut 55, 469–477 (2006).
Gianfrani, C. et al. Gliadin-specific type 1 regulatory T cells from the intestinal mucosa of treated celiac patients inhibit pathogenic T cells. J. Immunol. 177, 4178–4186 (2006).
van Leeuwen, M. A. et al. Macrophage-mediated gliadin degradation and concomitant IL-27 production drive IL-10- and IFN-gamma-secreting Tr1-like-cell differentiation in a murine model for gluten tolerance. Mucosal Immunol. 10, 635–649 (2017).
Huibregtse, I. L. et al. Induction of antigen-specific tolerance by oral administration of Lactococcus lactis delivered immunodominant DQ8-restricted gliadin peptide in sensitized nonobese diabetic Abo Dq8 transgenic mice. J. Immunol. 183, 2390–2396 (2009).
Girard-Madoux, M. J. et al. IL-10 control of CD11c + myeloid cells is essential to maintain immune homeostasis in the small and large intestine. Oncotarget 7, 32015–32030 (2016).
Shouval, D. S. et al. Interleukin-10 receptor signaling in innate immune cells regulates mucosal immune tolerance and anti-inflammatory macrophage function. Immunity 40, 706–719 (2014).
Zigmond, E. et al. Macrophage-restricted interleukin-10 receptor deficiency, but not IL-10 deficiency, causes severe spontaneous colitis. Immunity 40, 720–733 (2014).
Li, B. et al. IL-10 engages macrophages to shift Th17 cytokine dependency and pathogenicity during T-cell-mediated colitis. Nat. Commun. 6, 6131 (2015).
Jabri, B. & Abadie, V. IL-15 functions as a danger signal to regulate tissue-resident T cells and tissue destruction. Nat. Rev. Immunol. 15, 771–783 (2015).
Wapenaar, M. C. et al. The interferon gamma gene in celiac disease: augmented expression correlates with tissue damage but no evidence for genetic susceptibility. J. Autoimmun. 23, 183–190 (2004).
van Leeuwen, M. A. et al. Increased production of interleukin-21, but not interleukin-17A, in the small intestine characterizes pediatric celiac disease. Mucosal Immunol. 6, 1202–1213 (2013).
Sarra, M. et al. IL-15 positively regulates IL-21 production in celiac disease mucosa. Mucosal Immunol. 6, 244–255 (2013).
Bodd, M. et al. HLA-DQ2-restricted gluten-reactive T cells produce IL-21 but not IL-17 or IL-22. Mucosal Immunol. 3, 594–601 (2010).
Mucida, D. et al. Transcriptional reprogramming of mature CD4( + ) helper T cells generates distinct MHC class II-restricted cytotoxic T lymphocytes. Nat. Immunol. 14, 281–289 (2013).
Reimann, J. & Rudolphi, A. Co-expression of CD8 alpha in CD4 + T cell receptor alpha beta + T cells migrating into the murine small intestine epithelial layer. Eur. J. Immunol. 25, 1580–1588 (1995).
Reis, B. S., Rogoz, A., Costa-Pinto, F. A., Taniuchi, I. & Mucida, D. Mutual expression of the transcription factors Runx3 and ThPOK regulates intestinal CD4( + ) T cell immunity. Nat. Immunol. 14, 271–280 (2013).
Cortez, V. S. et al. CRTAM controls residency of gut CD4 + CD8 + T cells in the steady state and maintenance of gut CD4 + Th17 during parasitic infection. J. Exp. Med. 211, 623–633 (2014).
Takeuchi, A. et al. CRTAM determines the CD4 + cytotoxic T lymphocyte lineage. J. Exp. Med. 213, 123–138 (2016).
Matysiak-Budnik, T. et al. Alterations of the intestinal transport and processing of gliadin peptides in celiac disease. Gastroenterology 125, 696–707 (2003).
Cervantes-Barragan, L. et al. Lactobacillus reuteri induces gut intraepithelial CD4(+)CD8alphaalpha(+) T cells. Science 357, 806–810 (2017).
Jellison, E. R., Kim, S. K. & Welsh, R. M. Cutting edge: MHC class II-restricted killing in vivo during viral infection. J. Immunol. 174, 614–618 (2005).
Namekawa, T., Wagner, U. G., Goronzy, J. J. & Weyand, C. M. Functional subsets of CD4 T cells in rheumatoid synovitis. Arthritis Rheum. 41, 2108–2116 (1998).
Duftner, C. et al. Prevalence, clinical relevance and characterization of circulating cytotoxic CD4 + CD28- T cells in ankylosing spondylitis. Arthritis Res. Ther. 5, R292–R300 (2003).
Luda, K. M. et al. IRF8 transcription-factor-dependent classical dendritic cells are essential for intestinal T cell homeostasis. Immunity 44, 860–874 (2016).
Bouziat, R. et al. Reovirus infection triggers inflammatory responses to dietary antigens and development of celiac disease. Science 356, 44–50 (2017).
Acknowledgements
We thank B. Palanski (Stanford University) for providing the plasmid containing the TG2 mouse construct, S.M. Kim (University of Chicago) for anti-gluten and anti-TG2 ELISA protocols, B. Jabri (University of Chicago) for advice, F. Koning (University of Leiden) for critical reading of the manuscript, J. Brakshoofden and C. Gerkes (Erasmus Medical Center) for animal care. L.M.M.C. was funded by SSWO grants (SSWO 608, S16-23) and a research fellowship from the Dutch Sophia Research Foundation; S.V., a Dutch Sophia Research Foundation SSWO grant (S14−17); T.C., the Dutch Organization for Scientific research (NWO) (Innovative research incentives VIDI grant #91710377) and Genomics research initiative Zenith grant (#93512004). BEC received an NWO VIDI grant (#91776365).
Author information
Authors and Affiliations
Contributions
L.M.M.C. designed and performed experiments, analyzed data and wrote the manuscript. D.J.L.-K., L.Av.B., S.V., J.J.K., R.C.R. and Y.S.-O. performed experiments. M.G. and J.C.E. acquired patient material. B.E.C. and T.C. conceived the project. J.N.S. conceived the project, designed experiments, analyzed data, and wrote the manuscript. All authors provided comments on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Costes, L.M.M., Lindenbergh-Kortleve, D.J., van Berkel, L.A. et al. IL-10 signaling prevents gluten-dependent intraepithelial CD4+ cytotoxic T lymphocyte infiltration and epithelial damage in the small intestine. Mucosal Immunol 12, 479–490 (2019). https://doi.org/10.1038/s41385-018-0118-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41385-018-0118-0
This article is cited by
-
Extracellular CIRP induces CD4CD8αα intraepithelial lymphocyte cytotoxicity in sepsis
Molecular Medicine (2024)
-
IFN-γ+ cytotoxic CD4+ T lymphocytes are involved in the pathogenesis of colitis induced by IL-23 and the food colorant Red 40
Cellular & Molecular Immunology (2022)
-
A novel CD4+ CTL subtype characterized by chemotaxis and inflammation is involved in the pathogenesis of Graves’ orbitopathy
Cellular & Molecular Immunology (2021)