Abstract
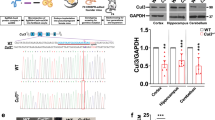
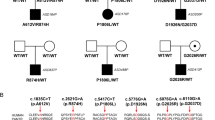
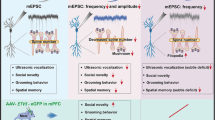
The pathophysiology of autism spectrum disorders (ASDs) is causally linked to postsynaptic scaffolding proteins, as evidenced by numerous large-scale genomic studies [1, 2] and in vitro and in vivo neurobiological studies of mutations in animal models [3, 4]. However, due to the distinct phenotypic and genetic heterogeneity observed in ASD patients, individual mutation genes account for only a small proportion (<2%) of cases [1, 5]. Recently, a human genetic study revealed a correlation between de novo variants in FERM domain-containing-5 (FRMD5) and neurodevelopmental abnormalities [6]. In this study, we demonstrate that deficiency of the scaffolding protein FRMD5 leads to neurodevelopmental dysfunction and ASD-like behavior in mice. FRMD5 deficiency results in morphological abnormalities in neurons and synaptic dysfunction in mice. Frmd5-deficient mice display learning and memory dysfunction, impaired social function, and increased repetitive stereotyped behavior. Mechanistically, tandem mass tag (TMT)-labeled quantitative proteomics revealed that FRMD5 deletion affects the distribution of synaptic proteins involved in the pathological process of ASD. Collectively, our findings delineate the critical role of FRMD5 in neurodevelopment and ASD pathophysiology, suggesting potential therapeutic implications for the treatment of ASD.

This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Data from this study are available from the authors upon reasonable request.
References
Lord C, Elsabbagh M, Baird G, Veenstra-Vanderweele J. Autism spectrum disorder. Lancet. 2018;392:508–20. https://doi.org/10.1016/S0140-6736(18)31129-2.
Lord C, Bishop SL. Recent advances in autism research as reflected in DSM-5 criteria for autism spectrum disorder. Annu Rev Clin Psychol. 2015;11:53–70. https://doi.org/10.1146/annurev-clinpsy-032814-112745.
Baribeau D, Anagnostou E. Novel treatments for autism spectrum disorder based on genomics and systems biology. Pharm Ther. 2022;230:107939 https://doi.org/10.1016/j.pharmthera.2021.107939.
Jiang C-C, Lin L-S, Long S, Ke X-Y, Fukunaga K, Lu Y-M, et al. Signalling pathways in autism spectrum disorder: mechanisms and therapeutic implications. Signal Transduct Target Ther. 2022;7:229 https://doi.org/10.1038/s41392-022-01081-0.
Shaw KA, McArthur D, Hughes MM, Bakian AV, Lee L-C, Pettygrove S, et al. Progress and disparities in early identification of autism spectrum disorder: autism and developmental disabilities monitoring network, 2002-2016. J Am Acad Child Adolesc Psychiatry. 2022;61:905–14. https://doi.org/10.1016/j.jaac.2021.11.019.
Chung C, Shin W, Kim E. Early and late corrections in mouse models of autism spectrum disorder. Biol Psychiatry. 2022;91:934–44. https://doi.org/10.1016/j.biopsych.2021.07.021.
Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–27.
Shcheglovitov A, Shcheglovitova O, Yazawa M, Portmann T, Shu R, Sebastiano V, et al. SHANK3 and IGF1 restore synaptic deficits in neurons from 22q13 deletion syndrome patients. Nature. 2013;503:267–71. https://doi.org/10.1038/nature12618.
Tatavarty V, Torrado Pacheco A, Groves Kuhnle C, Lin H, Koundinya P, Miska NJ et al. Autism-associated Shank3 is essential for homeostatic compensation in rodent V1. Neuron. 2020;106, https://doi.org/10.1016/j.neuron.2020.02.033.
Bourgeron T. From the genetic architecture to synaptic plasticity in autism spectrum disorder. Nat Rev Neurosci. 2015;16:551–63. https://doi.org/10.1038/nrn3992.
Wang T, Pei X, Zhan J, Hu J, Yu Y, Zhang H. FERM-containing protein FRMD5 is a p120-catenin interacting protein that regulates tumor progression. FEBS Lett. 2012;586:3044–50. https://doi.org/10.1016/j.febslet.2012.07.019.
Brázdová M, Quante T, Tögel L, Walter K, Loscher C, Tichý V, et al. Modulation of gene expression in U251 glioblastoma cells by binding of mutant p53 R273H to intronic and intergenic sequences. Nucleic Acids Res. 2009;37:1486–1500. https://doi.org/10.1093/nar/gkn1085.
Lu S, Ma M, Mao X, Bacino CA, Jankovic J, Sutton VR, et al. De novo variants in FRMD5 are associated with developmental delay, intellectual disability, ataxia, and abnormalities of eye movement. Am J Hum Genet. 2022;109:1932–43. https://doi.org/10.1016/j.ajhg.2022.09.005.
Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D, et al. DECIPHER: Database of chromosomal imbalance and phenotype in humans using ensembl resources. Am J Hum Genet. 2009;84:524–33. https://doi.org/10.1016/j.ajhg.2009.03.010.
Peca J, Feliciano C, Ting JT, Wang W, Wells MF, Venkatraman TN, et al. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437–42. https://doi.org/10.1038/nature09965.
Monteiro P, Feng G. SHANK proteins: roles at the synapse and in autism spectrum disorder. Nat Rev Neurosci. 2017;18:147–57. https://doi.org/10.1038/nrn.2016.183.
Lin YC, Frei JA, Kilander MB, Shen W, Blatt GJ. A subset of autism-associated genes regulate the structural stability of neurons. Front Cell Neurosci. 2016;10:263 https://doi.org/10.3389/fncel.2016.00263.
Groza T, Gomez FL, Mashhadi HH, Muñoz-Fuentes V, Gunes O, Wilson R, et al. The International Mouse Phenotyping Consortium: comprehensive knockout phenotyping underpinning the study of human disease. Nucleic Acids Res. 2022;51:D1038–D1045. https://doi.org/10.1093/nar/gkac972.
Iakoucheva LM, Muotri AR, Sebat J. Getting to the cores of autism. Cell. 2019;178:1287–98. https://doi.org/10.1016/j.cell.2019.07.037.
Quesnel-Vallières M, Weatheritt RJ, Cordes SP, Blencowe BJ. Autism spectrum disorder: insights into convergent mechanisms from transcriptomics. Nat Rev Genet. 2019;20:51–63. https://doi.org/10.1038/s41576-018-0066-2.
Mullins C, Fishell G, Tsien RW. Unifying views of autism spectrum disorders: a consideration of autoregulatory feedback loops. Neuron. 2016;89:1131–56. https://doi.org/10.1016/j.neuron.2016.02.017.
Moretto E, Murru L, Martano G, Sassone J, Passafaro M. Glutamatergic synapses in neurodevelopmental disorders. Prog Neuropsychopharmacol Biol Psychiatry. 2018;84:328–42. https://doi.org/10.1016/j.pnpbp.2017.09.014.
Lee E, Lee J, Kim E. Excitation/Inhibition imbalance in animal models of autism spectrum disorders. Biol Psychiatry. 2017;81:838–47. https://doi.org/10.1016/j.biopsych.2016.05.011.
Ecker C, Bookheimer SY, Murphy DG. Neuroimaging in autism spectrum disorder: brain structure and function across the lifespan. Lancet Neurol. 2015;14:1121–34. https://doi.org/10.1016/s1474-4422(15)00050-2.
Lewis JD, Evans AC, Pruett JR, Botteron K, Zwaigenbaum L, Estes A, et al. Network inefficiencies in autism spectrum disorder at 24 months. Transl Psychiatry. 2014;4:e388 https://doi.org/10.1038/tp.2014.24.
O’Reilly C, Lewis JD, Elsabbagh M. Is functional brain connectivity atypical in autism? A systematic review of EEG and MEG studies. PLoS One. 2017;12:e0175870 https://doi.org/10.1371/journal.pone.0175870.
Khoshbakht S, Beheshtian M, Fattahi Z, Bazazzadegan N, Parsimehr E, Fadaee M, et al. CEP104 and CEP290; genes with ciliary functions cause intellectual disability in multiple families. Arch Iran Med. 2021;24:364–73. https://doi.org/10.34172/aim.2021.53.
Silverman JL, Yang M, Lord C, Crawley JN. Behavioural phenotyping assays for mouse models of autism. Nat Rev Neurosci. 2010;11:490–502. https://doi.org/10.1038/nrn2851.
Orefice LL, Mosko JR, Morency DT, Wells MF, Tasnim A, Mozeika SM, et al. Targeting peripheral somatosensory neurons to improve tactile-related phenotypes in ASD models. Cell. 2019;178:867–86.e824. https://doi.org/10.1016/j.cell.2019.07.024.
Penagarikano O, Abrahams BS, Herman EI, Winden KD, Gdalyahu A, Dong H, et al. Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell. 2011;147:235–46. https://doi.org/10.1016/j.cell.2011.08.040.
Acknowledgements
This work was supported by the Ministry of Science and Technology of China (2021ZD0203204, 2022YFA1104003 and 2021YFC2501003) and grants from the National Natural Science Foundation of China (32030052, 31530028, 31720103908, 82230094, 81621063 and 81972616).
Author information
Authors and Affiliations
Contributions
YW and HQZ conceived and supervised the project. JD and JM performed the generation and breeding of the Frmd5 knock-out mice; TJL, JM, CL, DCY, XYZ, MYW performed the behavioral, molecular and biochemical experiments; GGX, CC and NYL performed electrophysiological and morphological experiments; TJL and CL performed the proteomics experiments; XYZ, TJL, JM and YNX wrote the manuscript. All of the authors read and accepted the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
All animal procedures performed in this study were approved by the Ethics Committee of Peking University Health Science Center (Beijing, China). For the data from the Decipher project, those who carried out the original analysis and collection of the data bear no responsibility for the further analysis or interpretation of the data.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lyu, TJ., Ma, J., Zhang, XY. et al. Deficiency of FRMD5 results in neurodevelopmental dysfunction and autistic-like behavior in mice. Mol Psychiatry (2024). https://doi.org/10.1038/s41380-024-02407-w
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41380-024-02407-w