Abstract
American Indians (AI) demonstrate the highest rates of both suicidal behaviors (SB) and alcohol use disorders (AUD) among all ethnic groups in the US. Rates of suicide and AUD vary substantially between tribal groups and across different geographical regions, underscoring a need to delineate more specific risk and resilience factors. Using data from over 740 AI living within eight contiguous reservations, we assessed genetic risk factors for SB by investigating: (1) possible genetic overlap with AUD, and (2) impacts of rare and low-frequency genomic variants. Suicidal behaviors included lifetime history of suicidal thoughts and acts, including verified suicide deaths, scored using a ranking variable for the SB phenotype (range 0–4). We identified five loci significantly associated with SB and AUD, two of which are intergenic and three intronic on genes AACSP1, ANK1, and FBXO11. Nonsynonymous rare and low-frequency mutations in four genes including SERPINF1 (PEDF), ZNF30, CD34, and SLC5A9, and non-intronic rare and low-frequency mutations in genes OPRD1, HSD17B3 and one lincRNA were significantly associated with SB. One identified pathway related to hypoxia-inducible factor (HIF) regulation, whose 83 nonsynonymous rare and low-frequency variants on 10 genes were significantly linked to SB as well. Four additional genes, and two pathways related to vasopressin-regulated water metabolism and cellular hexose transport, also were strongly associated with SB. This study represents the first investigation of genetic factors for SB in an American Indian population that has high risk for suicide. Our study suggests that bivariate association analysis between comorbid disorders can increase statistical power; and rare and low-frequency variant analysis in a high-risk population enabled by whole-genome sequencing has the potential to identify novel genetic factors. Although such findings may be population specific, rare functional mutations relating to PEDF and HIF regulation align with past reports and suggest a biological mechanism for suicide risk and a potential therapeutic target for intervention.
Similar content being viewed by others
Introduction
Suicide is a preventable public health problem that ranks as the second leading cause of death among young adults residing in the US [1]. Suicide rates remain even higher among American Indians/Alaska Natives (AI/AN), with rates more than 50% greater when compared to the general U.S. population across all age groups [2,3,4,5]. While suicide rates are unquestionably higher in the AI/AN population as a whole as compared to other US populations, rates also vary based on geographic region, tribal affiliation and whether one is living on a reservation [6,7,8,9,10,11,12].
American Indian/Alaska Natives also suffer from a disproportionate burden of the effects of alcohol, tobacco, and drug dependence [13, 14]. Large-scale U.S. epidemiological studies indicate that, compared with other U.S. ethnic groups, AI/AN demonstrate the highest rates of alcohol and other drug dependence [15, 16]. Additionally, lifetime rates of alcohol use disorders (AUD) differ depending on the tribes evaluated. Among individual tribal groups studied, reported rates of AUD have ranged from 20% to 70% [17,18,19,20]—significantly higher than epidemiological rates of AUD (14%) in the U.S. general population [21].
Substantial comorbidity has been demonstrated between SB and AUD. Chronic alcohol use is a clear risk factor for suicide [22], and having a severe use disorder and experiencing substance associated depression is particularly linked to increased risk for suicide [23]. Being acutely intoxicated provides additional risk for lethal suicide over and above the risk of chronic use [24, 25]. This is particularly true among American Indian/Alaska Natives [25, 26]. However, documenting comorbidity between disorders does not necessarily imply a causal connection between them or a common etiological pathway. Both AUD and suicide have been shown to have a significant genetic component to their etiology [27, 28]. Behavioral genetics studies have the advantage of being one of the strongest methods for determining whether the comorbidity among psychopathological conditions may be due to shared etiologies and/or pathologies associated with the disorders. For instance, a new study recently estimated the genetic correlation between suicide attempt and alcohol dependence at 44% in populations of European ancestry [29].
In recent years, numerous large genome-wide association studies (GWAS) have been reported for suicidal behaviors [28, 30,31,32,33,34]. These studies were carried out in populations dominated by individuals of European descent. Genetic epidemiology studies have estimated the heritability of SB ranging from 17% to 55% [35, 36]. Single nucleotide polymorphism (SNP)-heritability of suicide attempt using common genomic variants across the genome has been estimated to range from 3.5% in the UKBiobank [37] to 6.8% in the International Suicide Genetics Consortium (ISGC) [32]. The largest GWAS meta-analysis to date (with ~959 K individuals) reported 12 loci for suicide attempt. Risk loci were mostly intergenic and implicated genes DRD2, SLC6A9, FURIN, NLGN1, SOX5, PDE4B, and CACNG2 [30]. Intriguingly, one of the genes, ROBO2, reported by the ISGC GWAS to be associated with SB [31] is on a rare run-of-homozygosity (ROH) segment that was previously reported to be linked to severe AUD in an American Indian population [38].
While SNP-heritability and GWAS studies primarily estimate the impacts of common genomic variants, recent studies have shown that rare variants, especially those in low linkage disequilibrium (LD) with neighboring variants, are enriched for heritability for complex traits and diseases. Rare variants often represent recent and potentially deleterious mutations that can have biological consequences. Studies have indeed shown that most rare missense alleles in humans are deleterious [39]. In fact, a recent Utah Suicide Genetic Risk Study (USGRS) interrogating rare protein-coding variants included on the Illumina PsychArray chip has identified five rare variants for suicide death [40]. Genotyping chips, however, are usually not designed to capture rare variants in a population. Thus, to facilitate comprehensive studies of the impact of rare variants on a disease, exome sequencing or whole-genome sequencing is usually necessary.
The present report is part of a larger study exploring risk factors for substance dependence and suicide behaviors among American Indians [19, 41,42,43]. This American Indian (AI) population has been sequenced and deep phenotyped. The lifetime prevalence of AUD and suicide in this AI population is high, and evidence for heritability, linkage to specific chromosome locations, and genome-wide findings for AUD have been demonstrated [38, 42, 44,45,46,47]. The present study aimed to evaluate genetic factors associated with SB in this AI population. We hypothesized that certain genetic variants may underlie both SB and AUD, and further, that joint association analysis of related traits may lead to increased power to detect variants that contribute to both phenotypes. To this end, we conducted a bivariate genome association analysis between SB and AUD. We further hypothesized that rare (and low-frequency) variants in a high-risk population maybe a significant component of the complex genetic architecture underlying the disorder. Note that rare variants that occur in the general population might become less rare in population isolates such as American Indians. We thus conducted gene-based and pathway-based rare and low-frequency variant analyses for SB.
Material and Method
Participants
American Indian participants were recruited from eight geographically contiguous reservations. The reservations had a total population of about 3000 individuals. To be included in the study, participants had to be between the ages of 18 and 70 years, and mobile enough to be transported from their home to The Scripps Research Institute (TSRI). More details are given in Supplemental Materials. The demographics of the population is listed in Table S1. The protocol for the study was approved by the Institutional Review Board (IRB) of TSRI, and the board of the Indian Health Council, a tribal review group overseeing health issues for the reservations where the recruitment was undertaken. Written informed consent was obtained from each participant after the study was fully explained.
Phenotypes and genotypes
Potential participants first met individually with research staff, and during a screening period, participants completed a questionnaire that was used to gather information on demographics, personal medical history, and drinking history [48]. Each participant also completed an interview based on the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA) [49], which was used to collect lifetime history of two types of self-directed violence: suicidal thoughts including ideation (Have you ever thought about killing yourself?) and/or plans (Did you have a plan? Did you actually consider a way to take your life? What were you going to do?), as well as suicidal acts, including suicide attempt history (Have you ever tried to kill yourself? How did you try to kill yourself?) reported by the participants and suicide deaths obtained from community sources (e.g., verified by public records, family/tribal informants) over an 8-year period. From the lifetime history of suicidal thoughts and acts, we defined the suicidal behavior phenotype as a ranking variable: 0-none, 1-ideation, 2-plans, 3-attempts, 4-death. Since the individual risks are difficult to quantify, the scale as defined may not be the most ideal. However, the ranked numerical scale serves as an approximation to index corresponding risk as suicide risk increases in severity in this direction. In addition, it offers potential increase in statistical power for detection.
Diagnoses of lifetime DSM-5 AUD (mild, moderate, or severe) were also generated using the SSAGA. In addition, the interview retrospectively asks about the occurrence of alcohol-related life events, and the age at which the problem first occurred, from which a quantitative phenotype, the severity level of AUD, was derived. We used measures of the clinical course of alcoholism originally described by Schuckit and colleagues [50]. These measures were based on the relative order of the appearance of major “alcohol-related life events”. These life events have been shown to be highly similar and consistent across many different subgroups and populations, although age of onset and endorsement rates of individual events can differ [19, 51,52,53,54,55,56,57,58]. The severity level of AUD is indexed by the 36 alcohol-related life events (Table S2) in the clinical course of the disorder [19, 50], with life events given a severity weight of 1 for events 1-12; 2 for 13-24; and 3 for 25-36. AUD severity was then calculated as the sum of the severity weights of the 36 life events [46]. This metric gives larger weights to the more severe events that occur later in the clinical course and are more associated with severe use disorder [46, 53]. As shown in Supplementary Fig. S1, the AUD severity phenotype is highly aligned and correlated with the DSM-5 mild, moderate, and severe AUD diagnoses. Their Spearman’s rank correlation coefficient is 0.9. The relation between the SB phenotype and the AUD severity in this AI population is illustrated in Supplementary Fig. S2. The rank correlation between SB and AUD severity is 0.3.
Participants had low-coverage whole genome sequencing on blood-derived DNA using a previously described pipeline [59]. Briefly, the pair-end sequencing was performed on Illumina HiSeq2000 sequencers by a collaborating lab at the University of North Carolina at Chapel-Hill. About 80% of the samples had coverages between 3X and 12X. The qualities of the variant calling were confirmed using an Affymetrix Exome1A chip. Variants were removed if they had high missing rate ( > 5%), out of Hardy-Weinberg equilibrium (p < 1E-6), or had high Mendel error rate ( > 5%); individuals were removed if they had more than 2% genotypes missing. Further details can be found in the Supplementary Method.
Bivariate association analysis for suicidal behaviors and alcohol use disorders (SB-AUD)
We conducted genome-wide bivariate association analysis to identify genetic variants that might be associated with both SB and AUD severity (denoted as SB-AUD). By leveraging cross-trait covariance, multivariate tests for association may provide increased power over univariate tests. This occurs when the residual correlation between traits is opposite in direction to the genetic correlation induced by the genetic loci [60]. To control for population admixture and familial relatedness, we used the multivariate linear mixed model implemented as genome-wide efficient mixed-model association (GEMMA) for the study [61]. The bivariate association for each variant was conditioned on a genetic relationship matrix of the cohort derived from the genotypes, thus capturing a wide range of sample structures. Rare variants with minor allele frequency (MAF) lower than 1% were excluded from this analysis.
Gender, age, and age-squared were included as covariates, which we refer to as the main model. To take the social economic status of the AI cohort into consideration, in addition to the main model, we also tested an extended covariate model incorporating additional socioeconomic factors such as gross income, years of education, employment status, marriage status, religion, and frequency of religious service attendance. We first assessed whether the covariates were associated with SB or AUD severity through multiple regression prior to genetic association tests, and dropped the factors with negligible effects for both traits (p > 0.1). As a result, the extended model included years of education, employment, and religion as additional covariates (Supplemental Table S3A). Note that some of the socioeconomic factors in the extended model may also have genetic components.
Gene-based rare and low-frequency variant analysis for suicidal behaviors
Rare variants are usually tested by aggregating them into groups. We included both rare (MAF < 1%) and low-frequency (1% < = MAF < 5%) variants in this set of analyses. The variants were grouped by genes or by pathways and tested against the SB phenotype. 63% of all variants in the AI cohort are rare, while 12% are of low-frequency. We analyzed the rare and low-frequency variants across the genome using SKAT-O that optimally combines a burden test and a non-burden sequence kernel association test (SKAT) [62] within a linear mixed model as implemented in EPACTS [63]. We used recommended beta-density based weights beta(MAF;1,25), which increases the weights of rare variants while yielding decent nonzero weights for variants with MAF 1–5% [64]. In all sets of analyses, we used same main model and the extended model for covariates as described above but dropped the employment factor in the extended model (Supplemental Table S3B).
For each gene, we formed two types of groups. One group considered all variants on exons, 5’ and 3’ untranslated regions (UTRs), 50 base pairs (bp) upstream and 50 bp downstream of the gene (denoted as ExonReg). The other group included only the nonsynonymous variants and the splicing-site variants of the gene (denoted as Nonsyn). Intergenic and intronic variants were excluded in the present study. For each group type, a gene was excluded if fewer than three markers were found, or if less than 0.5% of the samples had any such markers on the gene. This resulted in 28,718 genes in the ExonReg group and 12,588 genes in the Nonsyn group. Association analysis was performed between each gene-based set and SB. False discovery rates (FDR) controlled by the Benjamini–Hochberg procedure (Benjamini and Hochberg, [65]) were used to set significant p values from the test statistics of the association tests. We combined two gene variant groups together for the multiple testing correction and report the FDR-adjusted p values (Yekutieli and Benjamini [66]). Note that the correction is conservative since the Nonsyn variants on a gene are a subset of the ExonReg variants, and thus they are correlated.
Pathway-based rare and low-frequency variant analysis for suicidal behaviors
We utilized a collection of canonical pathways from the Molecular Signatures Database (MSigDB) version 7.5.1 [67, 68]. There are total of 2981 pathway gene sets in this release. Since a pathway may contain a large number of genes, to limit the number of variants included in each set for test, we only considered rare and low-frequency nonsynonymous and splicing-site variants (Nonsyn) on the genes within each pathway. The same filters as in the gene-based tests were applied. We performed SKAT-O tests on the Nonsyn variant group for each pathway to be associated with SB. FDR-adjusted p-values are reported.
Rare and low-frequency variant analysis for SB-AUD
We carried out the same set of analyses as described in the last two sections for AUD severity as well. We then used meta-analysis to obtain the effects of rare and low-frequency variants on SB-AUD [69]. Since the test statistics for the two traits were correlated, we corrected these correlations prior to meta-analysis [70]. Details are given in Supplemental Method.
Functional analysis
For the top variants identified in the bivariate association analysis and for the top genes identified in the rare and low-frequency variant analysis, we obtained the combined annotation dependent depletion (CADD v1.6) scores [71] for the variants included in the test to assess the deleteriousness of these variants. CADD integrates multiple functional annotations and genome-wide variant effect prediction models to produce a scaled C-score. We report the number of variants tested in each gene that have C-scores in the range of 15–20 (1–3% most deleterious mutations), 20–30 (0.1–1%), and over 30 ( < 0.1%). We chose 15 as the lowest C-score to report as it happened to be the median value for all possible canonical splice site changes and nonsynonymous variants in CADD v1.0 [72].
The variants with p-value < 10−6 from the bivariate analysis were annotated with the nearest genes, and the associated set of genes was then subjected to functional enrichment analysis using GENE2FUNC in FUMA version 1.5.1 [73]. An integrated network analysis was completed using GeneMANIA [74]. The same set of functional analysis was applied to the top genes from the rare and low-frequency variants analysis as well.
More details can be found in Supplemental Method.
Results
A total of 743 participants have sequencing data and a non-missing phenotype for SB (none [0], suicidal ideation [1], suicide planning [2], suicide attempt [3], suicide death [4], and 742 participants have data on AUD severity (range 0–69, mean = 22.6, sd = 18.8). 57.3% of these individuals are female. Their average age is 31. The detailed demographics is listed in Table S1. The sample size in the main covariate model is 742 for the SB-AUD bivariate analysis and 743 for the SB rare and low-frequency variant analysis, and reduces to 720 and 740 respectively in the extended covariate model due to missing data (Table S3).
Bivariate genome-wide significant variants associated with SB-AUD in AI
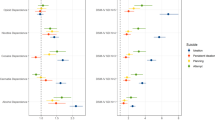
Five variants were identified as being significantly associated with AUD severity and SB (p < 5E-8) using the main covariate model, as illustrated in Fig. 1. Only one of the variants, rs184204326 on gene FBXO11, has MAF over 5% (Table 1). Although the association of this variant is mostly driven by SB (p = 1.48E-07), the bivariate association is statistically more significant (p = 3.63E-08) than the univariate associations, which also holds for three other top variants, including an intergenic variant between genes ZIC2 and PCCA, variant rs76300969 on gene AACSL, and variant rs530542541 on ANK1. Among the top variants shown in Table 1, rs184204326 is the only SNP for which brain expression quantitative trait loci (eQTL) were found in the Braineac database [75]. That variant is associated with the differential gene expression of several genes in brain tissues including: putamen, substantia nigra, thalamus, occipital cortex, and temporal cortex (Table S4). The most significant eQTL is for EPCAM (encoding epithelial cell adhesion molecule) expression in the putamen tissue (p = 0.0019). The variant is also associated with gene expression in several brain tissues for FBXO11, MCFD2 (encoding a soluble luminal protein), and KCNK12 (encoding a potassium channel protein). One variant on LRRC4C was significantly associated with AUD severity and SB in the extended covariate model (Table S5). The gene has been associated with drinking and smoking behaviors [76] as well as educational attainment [77]. Four of the five significant variants in the main model remained suggestive significant in the extended model; one was dropped in the analysis due to low allele frequency. As a comparison, we also conducted an additional set of bivariate analysis using SB and the DSM5 AUD diagnosis phenotype (Table S5). While DSM5 AUD diagnosis and the clinical course-derived AUD severity phenotypes are highly correlated (Fig. S1), the latter offers increased power for detection as shown in Table S5.
Variants associated with SB-AUD at p < 10−6 (using the main model) were mapped to 31 genes. The majority of these variants are intronic, upstream, or downstream from a gene. Rs1804145 on SERPINF1 is the only nonsynonymous variant and has a C-score of 22.8 (top 0.52% most deleterious). These genes are enriched for 27 transcription factor (TF) targets and three microRNA targets (Table S6), suggesting that the top genes share certain regulatory motifs. These genes are most significantly up-regulated in artery, and down-regulated in areas included in the basal ganglia (nucleus accumbens, caudate, and putamen). In addition, they are significantly differentially expressed in anterior cingulate cortex, hypothalamus, substantia nigra, and kidney (Fig. 2). Many psychiatric or neurological disorders have been linked to basal ganglia targets including addiction and depression [78].
The tissue-specific differential gene expression test was conducted against all genes across genomes that exhibited significantly increased or decreased expression levels in a certain tissue sample compared to all other samples. The analysis was performed using FUMA and utilized tissue-specific transcriptome data across 54 tissue types from GTEx v8. From top to bottom: up-regulated, down-regulated, differentially expressed. Red: Significantly enriched (p < 0.05 with Bonferroni correction).
Genes with rare and low-frequency variants associated with suicidal behaviors in AI
Rare and low-frequency variant analysis identified six genes and one long intergenic non-coding RNA (lincRNA) as being significantly associated with SB (FDR adjusted p < 0.05) using the main model, and an additional four genes strongly associated with SB (FDR < 0.1) (Table 2). SERPINF1 was the top gene linked to SB. Both the nonsyn (nominal p = 1.3E-6) and the exonreg (nominal p = 1.7E-6) variant groups of this gene remained genome wide significant after the multiple testing correction (FDR = 0.024). Four out of the nine nonsyn rare and low-frequency variants in SERPINF1 had C-scores between 23 and 29, representing between 0.1 and 0.5% most deleterious mutations. One of the four probably damaging variants, rs1804145, was also suggestively associated with SB-AUD in the bivariate analysis (p = 1.1E-7, see Table 1). Additional genes significantly associated with SB included: ZNF30, CD34, SLC5A9, OPRD1, and HSD17B3. A lincRNA AC002511.3 is also associated with SB. Of the 15 nonsyn variants on ZNF30, one has a C-score of 33 (0.05% most deleterious), and six have C-scores between 22 and 24 ( ~ 0.5% most deleterious). One of the nine nonsyn variants on CD34 has a C-score of 32. Seven out of the 12 nonsyn variants on SCL5A9 have C-scores ranging from 24 to 30. OPRD1 has three rare exon variants, one of which has a C-score of 18 (1.6% most deleterious), and one of the 10 rare variants on HSD17B3 has a C-score of 22.8 ( ~ 0.5% most deleterious). Using the extended model as shown in Table S7, four genes and one lincRNA had FDR < 0.05 and four genes had FDR < 0.1, including all genes that were significantly associated with SB in the main model.
A gene set enrichment analysis for the 11 genes whose rare and low-frequency variants were associated with SB at FDR < 0.1 using the main model (Table 2) identified axon to be the most significantly enriched gene ontology term (nominal p = 1.63E-5, adjusted p = 0.016). Four of the 11 genes belong to the axon gene set: OPRD1, NRP1, SERPINF1, and DCTN1. Network analysis conducted using GeneMania for the 11 genes in Table 2 recognized 10 genes and automatically selected 10 additional related genes (Fig. 3). The analysis considered pathways and genetic interactions and identified a number of significantly associated functional networks (Table S9). Figure 3 illustrates the top enriched distinct functions, including VEGF signaling (FDR = 2.1E-04), angiogenesis (FDR = 2.1E-04), and response to decreased oxygen levels (FDR = 4.2E-03). VEGF, as an angiogenic cytokine, and hypoxia have been associated with depression and suicide [79, 80].
Three of the enriched functional networks are highlighted in the gene nodes (see Table S9 for the complete list). Each circle represents a gene and the color on the circle indicates to which pathway the gene belongs to. Yellow: vascular endothelial growth factor (VEGF) receptor signaling pathway (FDR = 2.1E-04); Red: angiogenesis (FDR = 2.1E-04); Blue: response to decreased oxygen levels (FDR = 4.2E-03). The color of the edges linking the genes indicate the type of database used in the GeneMania analysis. Blue: Pathways; Green: Genetic interactions.
From the meta-analysis between SB and AUD severity for the rare and low-frequency variant analysis, none of the nominally significant genes or pathways remained genome-wide significant after multiple comparison corrections (Table S10).
Pathways with rare and low-frequency variants associated with suicidal behaviors in AI
Regulation of gene expression by hypoxia inducible factor (HIF) is the top pathway (nominal p = 1.4E-5, FDR = 0.04) whose rare and low-frequency variants were associated with SB using the main model (Table 3). The pathway is comprised of 10 genes with total of 91 nonsynonymous or splicing site variants (nonsyn), of which 83 have MAF < 5% and were included in the association test. Hypoxia could decrease serotonin synthesis, which has been linked to suicide [81]. The three additional pathways that were associated with SB at FDR < 0.1 using the main model are essentially two distinct pathways: vasopressin regulated water reabsorption with 164 nonsyn rare and low-frequency variants on 44 genes, and cellular hexose transport with 126 nonsyn rare and low-frequency variants on 21 genes. Vasopressin plays a critical role in regulating renal water reabsorption and cardiovascular homeostasis. As an integral part of the hypothalamic-pituitary-adrenal (HPA) axis, it is also an important player in response to stress and contributes to stress-related disorders such as anxiety and depression [82]. The cellular hexose transport pathway includes gene families responsible for glucose transport in humans. Sodium independent glucose transporters (GLUTs) are encoded by the SLC2 family, while sodium dependent glucose transporters (SGLTs) are encoded by the SLC5 family. Using the extended model, as shown in Table S8, all four pathways were significantly associated with SB (FDR = 0.019–0.024).
Overall, there was no major discrepancy between the findings from the main covariate model and the extended model although extended model had reduced sample sizes and the significant levels differed. However, some of the social economic factors included in the extended model also have genetic components. For instance, it has been demonstrated that suicide death and suicide behavior as well as AUD showed negative genetic correlation with educational attainment [31, 83]. Thus, it is not clear how educational attainment might impact the genetic associations when included as a covariate: more detailed analysis is beyond the scope of this study. We will base our discussions on the findings from the main model.
Discussion
The present study investigated potential genetic risk factors for suicidal behaviors in an American Indian population living on reservations that demonstrates high rates of suicide as well as AUD [19, 43]. We identified five variants significantly associated with SB-AUD, two of which are intergenic and three are intronic. Six genes, one lincRNA, and one canonical pathway, were found to be significantly associated with SB through rare and low-frequency variant analysis. Four additional genes, and two pathways, also were strongly associated with SB in this AI cohort. Approximately 3–20% (50–79%) individuals carry some of the rare and low-frequency variants on these genes (pathways).
F-box gene FBXO11 significantly associated with SB-AUD in this AI population
The intronic variants significantly associated with SB-AUD are on genes AACSP1, ANK1, and FBXO11. AACSP1 has been associated with an Alzheimer’s marker through interaction [84]. ANK1 has also been linked to Alzheimer’s disease through epigenetic deregulation [85, 86] and appears to play a role in immunomodulation [87]. Rs184204326 on gene FBXO11 was the only common variant significantly associated with SB-AUD. FBXO11 is highly conserved in evolution, and involved in the maintenance of genome stability [88] and the regulation of alternative splicing [89]. While the gene is expressed in various tissues, it is particularly abundant in the brain. FBXO11 has not been previously associated with SB. However, variants on or near FBXO11 have been associated with a number or correlated behavior traits and disorders such as alcohol consumption [76], smoking initiation [76, 90], risk taking behaviors [91, 92], externalizing behaviors [93], educational attainment [77], insomnia [94], schizophrenia and depression [95, 96]. De novo mutations on FBXO11 were found to cause intellectual disability with behavior problems as well as facial dysmorphisms [97] and other neurodevelopmental disorders [98]. Variant rs77969729 on FBXO11 has been linked to Alzheimer’s disease in the UKBiobank [99]. This variant is in LD with rs184204326 on FBXO11 in the AI cohort and associated with SB-AUD at p = 3.4E-7.
Rare and low-frequency mutations in PEDF are likely a risk factor for suicidal behaviors
SERPINF1 was the top gene significantly associated with SB in the rare and low-frequency variant analysis. It encodes serpin F1, and is also known as pigment epithelium-derived factor (PEDF). This gene has nine rare and low-frequency nonsynonymous or splicing site mutations, of which four are among the 0.1–0.5% most deleterious. Nearly 6% of individuals in the AI cohort carry some of these mutations. PEDF is a secreted glycoprotein and a potent inhibitor of angiogenesis [100]. It has been linked to obesity [101]. It also has neuroprotective effects and been implicated in depression [102, 103]. Reduction in PEDF levels have been found in the plasmas of patients with major depressive disorder (MDD), as well as in the prefrontal cortex (PFC) of animal models exhibiting depressive-like behaviors [103]. Conversely, overexpression of PEDF in the PFC has been shown to induce antidepressant “like” behaviors, by exerting effects on the tryptophan and glutamate in the PFC, with tryptophan being an essential amino acid precursor of serotonin, which is known to be associated with both depression and SB [104]. Another study has shown that PEDF in the hippocampus has a similar effect on depressive phenotypes in animal models by contributing to the synaptic formation and Wnt signaling activation in that region [105]. PEDF has thus been suggested as a biomarker and a novel therapeutic target for depression [102].
Additional genes significantly associated with SB included ZNF30, CD34, SLC5A9, OPRD1, and HSD17B3. ZNF30 encodes a zinc finger protein. A microdeletion of five genes including ZNF30 results in chromosome 19q13.11 deletion syndrome that includes features such as: developmental delay, microcephaly and intellectual disabilities [106]. The GWAS meta-analysis by the international suicide genetics consortium (ISGC) reported another zinc finger family gene (ZNF28) associated with SB and suicide death [31]. CD34 is involved in the innate immune system. Its variants have been associated with Alzheimer’s disease [99] and risk-taking behavior [92]. SCL5A9 is involved in sodium ion transport and is also known to be a sodium-dependent glucose transporter [107]. This gene has been associated with amygdala volume [108] and metabolic measurements [109]. Variants on or near HSD17B3 have been linked to smoking behavior [110], brain morphology measurements [111], memory performance [112] and Alzheimer’s markers [84, 113].
Opioid receptor delta 1 (OPRD1) gene encodes a member of the opioid family of the G-protein coupled receptors (GPCR). Delta opioid receptors are generally involved in reward mediation and neuroprotection. OPRD1 is specifically involved in the opioid receptor signaling pathway and cellular response to hypoxia. Numerous candidate gene studies have implicated OPRD1 in addiction, including opioid, cocaine, and alcohol dependence [114, 115]. Variants on OPRD1 have also been associated with schizophrenia [116] and educational attainment [77].
Hypoxia regulation is significantly associated with suicidal behaviors
Nonsyn rare and low-frequency variants in a pathway related to hypoxia inducible factor (HIF) regulation were significantly associated with SB (see Table 3). Nearly half of the individuals in the AI cohort have some of these mutations on their genes in this pathway. The network analysis of the top rare and low-frequency variant genes associated with SB also found that those genes were enriched for response to decreased oxygen levels (see Fig. 3).
Chronic hypoxia is suggested to be a risk factor for suicide [79], and metabolic stress associated with hypoxia is a possible mechanism [117]. Hypoxia is also hypothesized to increase the risk of suicide by reducing the synthesis of brain serotonin [81] or downregulating PEDF [118], which has a protective role in depression and is associated with SB in this AI cohort. HIFs are transcriptional factors that respond to reduced oxygen levels in cell and tissue. HIF-1 protects against hypoxia and reduces oxidative stress [119], while HIF-2α plays an important role in the modulation of inflammatory responses [120]. Recent studies have indicated that oxidative stress and abnormal energy metabolism in the brain play significant roles in the development of depression. Therefore, increasing HIF-1 activity has been suggested as a potential new therapeutic target for depression and suicide ideation [119]. Gene expression analyses have demonstrated that patients with MDD exhibit increased expression of HIF-1 and its target genes, including VEGF and GLUT1 (SLC2A1) [80]. Our network analysis of the top rare and low-frequency variant genes associated with SB in the AI have also found enrichments in VEGF receptors signaling and angiogenesis.
Alcohol exposure also can alter expression of HIF. For instance, alcohol exposure can induce HIF-1α activation, however the dose and timing of alcohol exposure results in differential expression of HIF-1α in the brain and other organs [121]. Both acute and chronic alcohol exposure have been found to increase HIF-1α expression in the brain cortex, whereas chronic binge alcohol exposure decreased HIF-1α expression [121]. Another study found that HIF3A could be epigenetically induced in the amygdala in animal models by acute alcohol exposure, and its epigenetic reprogramming was associated with the anti-anxiety effect of acute alcohol exposure [122]. Nonsyn rare and low-frequency variants in HIF3A were strongly associated with SB (nominal p = 2.9E-05, FDR = 0.09) in the present study.
Vasopressin-regulated water metabolism and hexose transport strongly associated with suicidal behaviors
Nonsyn rare and low-frequency variants in two additional pathways, vasopressin-regulated water reabsorption and cellular hexose transport, were strongly associated with SB in the AI. Vasopressin is an evolutionary ancient neuropeptide involved in regulating physiological processes such as renal water reabsorption and cardiovascular homeostasis. It also plays an important role in the modulation of emotional and social behaviors in the brain [123, 124], with vasopressin containing neurons most abundantly found in the hypothalamus. The vasopressin system is known to interact with the HPA axis [82]. Indeed, cortisol response and stress reactivity within the HPA axis is well-established as an endophenotype for depression and SB, as increased cortisol level is associated with death by suicide [125, 126]. HPA axis dysfunction has also been observed in those with a history of suicide attempts [126]. In addition, changes in water and electrolyte metabolism have been reported in clinical studies of depressed patients [127, 128]. Alcohol also uniquely interacts with the vasopressin system. Alcohol is a diuretic that promotes water loss by inhibiting the production of vasopressin. The HPA axis response to alcohol can be altered by manipulating the vasopressin system [129]. The vasopressin system has thus become an emerging therapeutic target for stress and depression, as well as alcohol-related behaviors [82, 129]. Gene families in the cellular hexose transport pathway mediate glucose absorption in the small intestine, glucose reabsorption in the kidney, glucose uptake by the brain across the blood-brain barrier, and glucose release by all cells in the body [130]. Evidence suggests that disturbances in glucose metabolism may be associated with suicidal ideation and attempts [131]. A recent study has found a significant association between blood glucose and suicide attempts in male patients with MDD [132]. Taken together, our findings suggest that pathways underlying vasopressin-regulated water reabsorption and the cellular hexose transport system warrant additional investigation in association with SB.
Strengths and limitations
The results of this study should be interpreted in light of several limitations. The analyses were not meant to generate a comprehensive model of suicide risk and AUD in this community group, but rather to determine whether specific genetic associations could be identified for suicidal thoughts and acts, and between SB and AUD phenotypes. A larger sample, powered to access and assess additional variables associated with suicide risk, particularly according to the above systems, is recommended. We further emphasize that American Indians are a diverse group both genetically and environmentally, therefore our findings may not generalize to other American Indians in the population from which the sample was drawn or be representative of all American Indians, as rates of AUD and suicide vary among tribes [17, 19, 133]. The population diversity in fact underscores the need to delineate risk and resilience factors in local communities to develop community-specific prevention and intervention efforts [10,11,12, 134]. Since our study utilized both whole genome sequence data and an extensive phenotyping instrument, there is presently no replication AI sample available to our knowledge, especially given that suicide risks in American Indians are overall understudied. While our specific findings may not generalize to other populations, the identified genes and functional pathways for SB may be shared by different populations thus warrant further investigations.
We used retrospective data for lifetime measures of suicide risk, which are subject to recall bias and may include reporting bias for psychiatric symptoms. Lifetime suicidal ideation and behaviors were assessed using a subscale of the SSAGA as well as verified death records. Suicidal behaviors were defined using a ranked numerical scale to index corresponding risk. Although suicide risk increases in severity in this direction, suicidal behaviors range in severity by outcomes of risk (e.g., suicidal ideation to suicide attempts to death by suicide) and within such outcomes (i.e., according to intensity, duration, and pervasiveness of suicidal ideation; the lethality of a suicide attempt, etc.). Future studies using validated measures of suicidal behavior, such as the Columbia-suicide severity rating scale (CSSRS), are warranted to increase specificity and precision within this area.
In conclusion, we conducted the first genome-wide bivariate association analysis for SB and AUD, and identified five genome-wide significant loci. We also conducted the first large-scale rare and low-frequency variant analysis and identified 11 novel genes and three pathways for SB. Of particular importance, this study represents the first investigation of genetic factors for SB in an American Indian population that has high risk for suicide. Although our findings may be population specific, the rare and low-frequency functional mutations relating to PEDF and HIF regulation may suggest an important mechanism underlying suicidal behaviors and a potential therapeutic target for treatment and intervention in the prevention of suicide.
Data availability
The data that support the finding of this study are available by contacting the last author. However, data availability is subject to approval of the specific AI tribes participating. The canonical pathways as gene symbols were downloaded from GSEA (http://www.gsea-msigdb.org/gsea/msigdb/download_file.jsp?filePath = /msigdb/release/7.5.1/c2.cp.v7.5.1.symbols.gmt). The hg19 gene list was downloaded from: https://yanglab.westlake.edu.cn/software/gcta/res/glist-hg19.txt. GEMMA version 0.95alpha and EPACTS version 3.4.2 were used and can be downloaded from github.
References
Curtin SC, Warner M, Hedegaard H. Increase in Suicide in the United States, 1999-2014. NCHS. Data Brief. 2016;241:1–8.
Department of Health and Human Services, Indian Health Service, Office of Public Health Support, Division of Program Statistics. Trends in Indian Health 2014. 2014: [Available from: https://www.ihs.gov/dps/publications/trends2014/.
Karaye IM. Differential trends in US suicide rates, 1999–2020: Emerging racial and ethnic disparities. Preventive Med. 2022;159:107064.
Leavitt R, Ertl A, Sheats K, Petrosky E, Ivey-Stephenson A, Fowler K. Suicides Among American Indian/Alaska Natives — National Violent Death Reporting System, 18 States, 2003–2014. MMWR Morb Mortal Wkly Rep. 2018;67:237–42.
Wilson RF, Liu G, Lyons BH, Petrosky E, Harrison DD, Betz CJ, et al. Surveillance for Violent Deaths — National Violent Death Reporting System, 42 States, the District of Columbia, and Puerto Rico, 2019. MMWR Surveill Summaries. 2022;71:1–40.
Bolton S-L, Elias B, Enns MW, Sareen J, Beals J, Novins DK. A comparison of the prevalence and risk factors of suicidal ideation and suicide attempts in two American Indian population samples and in a general population sample. Transcultural. Psychiatry 2013;51:3–22.
Chachamovich E, Kirmayer LJ, Haggarty JM, Cargo M, McCormick R, Turecki G. Suicide among Inuit: Results from a Large, Epidemiologically Representative Follow-Back Study in Nunavut. Can J Psychiatry. 2015;60:268–75.
Cwik M, Doty SB, Hinton A, Goklish N, Ivanich J, Hill K, et al. Community Perspectives on Social Influences on Suicide Within a Native American Reservation. Qualitative Health Res. 2021;32:16–30.
Freedenthal S, Stiffman AR. Suicidal Behavior in Urban American Indian Adolescents: A Comparison with Reservation Youth in a Southwestern State. Suicide Life-Threatening Behav. 2004;34:160–71.
Livingston R, Daily RS, Guerrero APS, Walkup JT, Novins DK. No Indians to Spare: Depression and Suicide in Indigenous American Children and Youth. Child Adolesc Psychiatr Clin North Am. 2019;28:497–507.
Shaw JL, Beans JA, Comtois KA, Hiratsuka VY. Lived Experiences of Suicide Risk and Resilience among Alaska Native and American Indian People. Int J Eviron Res Public Health. 2019;16:3953.
Trout L, Wexler L. Arctic Suicide, Social Medicine, and the Purview of Care in Global Mental Health. Health Hum Rights. 2020;22:77–89.
Indian Health Service. U S Indian Health Service, Division of Program Statistics: Trends in Indian Health. Rockville, MD: U S Department of Health and Human Services, Public Health Service, Indian Health Service; 1997.
Spillane NS, Kirk-Provencher KT, Schick MR, Nalven T, Goldstein SC, Kahler CW. Identifying Competing Life Reinforcers for Substance Use in First Nation Adolescents. Subst Use Misuse. 2020;55:886–95.
Compton WM, Thomas YF, Stinson FS, Grant BF. Prevalence, correlates, disability, and comorbidity of DSM-IV drug abuse and dependence in the United States: results from the national epidemiologic survey on alcohol and related conditions. Arch Gen Psychiatry. 2007;64:566–76.
Brave Heart MYH, Lewis-Fernández R, Beals J, Hasin DS, Sugaya L, Wang S, et al. Psychiatric disorders and mental health treatment in American Indians and Alaska Natives: results of the National Epidemiologic Survey on Alcohol and Related Conditions. Soc Psychiatry Psychiatr Epidemiol. 2016;51:1033–46.
Beals J, Novins DK, Whitesell NR, Spicer P, Mitchell CM, Manson SM. Prevalence of Mental Disorders and Utilization of Mental Health Services in Two American Indian Reservation Populations: Mental Health Disparities in a National Context. Am J Psychiatry. 2005;162:1723–32.
Spicer P, Beals J, Croy CD, Mitchell CM, Novins DK, Moore L, et al. The prevalence of DSM-III-R alcohol dependence in two American Indian populations. Alcohol Clin Exp Res. 2003;27:1785–97.
Ehlers CL, Wall TL, Betancourt M, Gilder DA. The clinical course of alcoholism in 243 Mission Indians. Am J Psychiatry. 2004;161:1204–10.
Robin RW, Long JC, Rasmussen JK, Albaugh B, Goldman D. Relationship of Binge Drinking to Alcohol Dependence, Other Psychiatric Disorders, and Behavioral Problems in an American Indian Tribe. Alcohol: Clin Exp Res. 1998;22:518–23.
Grant BF, Goldstein RB, Saha TD, Chou SP, Jung J, Zhang H, et al. Epidemiology of DSM-5 alcohol use disorder: Results from the national epidemiologic survey on alcohol and related conditions III. JAMA Psychiatry. 2015;72:757–66.
Kaplan MS, Huguet N, McFarland BH, Caetano R, Conner KR, Nolte KB, et al. Heavy Alcohol Use Among Suicide Decedents: Differences in Risk Across Racial-Ethnic Groups. Psychiatr Serv. 2016;67:258.
Ehlers CL, Kim C, Gilder DA, Stouffer GM, Caetano R, Yehuda R. Lifetime history of traumatic events in a young adult Mexican American sample: Relation to substance dependence, affective disorder, acculturation stress, and PTSD. J Psychiatr Res. 2016;83:79–85.
Cherpitel CJ, Borges GLG, Wilcox HC. Acute Alcohol Use and Suicidal Behavior: A Review of the Literature. Alcohol: Clin Exp Res. 2004;28:18S–28S.
Kaplan MS, Giesbrecht N, Caetano R, Conner KR, Huguet N, McFarland BH, et al. Acute Alcohol Consumption as a Contributing Factor to Suicidal Behavior. Am J Public Health. 2013;103:e2–e3.
Caetano R, Kaplan MS, Kerr W, McFarland BH, Giesbrecht N, Kaplan Z. Suicide, Alcohol Intoxication, and Age Among Whites and American Indians/Alaskan Natives. Alcohol: Clin Exp Res. 2020;44:492–500.
Edenberg HJ, Gelernter J, Agrawal A. Genetics of Alcoholism. Curr Psychiatry Rep. 2019;21:26.
Erlangsen A, Appadurai V, Wang Y, Turecki G, Mors O, Werge T, et al. Genetics of suicide attempts in individuals with and without mental disorders: a population-based genome-wide association study. Mol Psychiatry. 2020;25:2410–21.
Stephenson M, Lannoy S, Edwards AC. Shared genetic liability for alcohol consumption, alcohol problems, and suicide attempt: Evaluating the role of impulsivity. Transl Psychiatry. 2023;13:87.
Docherty AR, Mullins N, Ashley-Koch AE, Qin XJ, Coleman J, Shabalin AA, et al. Genome-wide association study meta-analysis of suicide attempt in 43,871 cases identifies twelve genome-wide significant loci. medRxiv. 2022:2022.07.03.22277199.
Li QS, Shabalin AA, DiBlasi E, Gopal S, Canuso CM, Palotie A, et al. Genome-wide association study meta-analysis of suicide death and suicidal behavior. Mol Psychiatry. 2023;28:891–900.
Mullins N, Kang J, Campos AI, Coleman JRI, Edwards AC, Galfalvy H, et al. Dissecting the Shared Genetic Architecture of Suicide Attempt, Psychiatric Disorders, and Known Risk Factors. Biol Psychiatry. 2022;91:313–27.
Kimbrel NA, Ashley-Koch AE, Qin XJ, Lindquist JH, Garrett ME, Dennis MF, et al. A genome-wide association study of suicide attempts in the million veterans program identifies evidence of pan-ancestry and ancestry-specific risk loci. Mol Psychiatry. 2022;27:2264–72.
Strawbridge RJ, Ward J, Ferguson A, Graham N, Shaw RJ, Cullen B, et al. Identification of novel genome-wide associations for suicidality in UK Biobank, genetic correlation with psychiatric disorders and polygenic association with completed suicide. eBioMedicine. 2019;41:517–25.
Fu Q, Heath AC, Bucholz KK, Nelson EC, Glowinski AL, Goldberg J, et al. A twin study of genetic and environmental influences on suicidality in men. Psychol Med. 2002;32:11–24.
Voracek M, Loibl LM. Genetics of suicide: a systematic review of twin studies. Wien klinische Wochenschr. 2007;119:463–75.
Ruderfer DM, Walsh CG, Aguirre MW, Tanigawa Y, Ribeiro JD, Franklin JC, et al. Significant shared heritability underlies suicide attempt and clinically predicted probability of attempting suicide. Mol Psychiatry. 2020;25:2422–30.
Peng Q, Ehlers CL. Long tracks of homozygosity predict the severity of alcohol use disorders in an American Indian population. Mol Psychiatry. 2021;26:2200–11.
Kryukov GV, Pennacchio LA, Sunyaev SR. Most Rare Missense Alleles Are Deleterious in Humans: Implications for Complex Disease and Association Studies. Am J Hum Genet. 2007;80:727–39.
DiBlasi E, Shabalin AA, Monson ET, Keeshin BR, Bakian AV, Kirby AV, et al. Rare protein-coding variants implicate genes involved in risk of suicide death. Am J Med Genet Part B: Neuropsychiatr Genet. 2021;186:508–20.
Ehlers CL, Gilder DA, Gizer IR, Wilhelmsen KC. Indexing the ‘dark side of addiction’: substance-induced affective symptoms and alcohol use disorders. Addiction. 2019;114:139–49.
Ehlers CL, Gizer IR. Evidence for a genetic component for substance dependence in Native Americans. Am J Psychiatry. 2013;170:154–64.
Ehlers CL, Yehuda R, Gilder DA, Bernert R, Karriker-Jaffe KJ. Trauma, historical trauma, PTSD and suicide in an American Indian community sample. J Psychiatr Res. 2022;156:214–20.
Peng Q, Gizer IR, Libiger O, Bizon C, Wilhelmsen KC, Schork NJ, et al. Association and ancestry analysis of sequence variants in ADH and ALDH using alcohol-related phenotypes in a Native American community sample. Am J Med Genet Part B: Neuropsychiatr Genet. 2014;165:673–83.
Peng Q, Gizer IR, Wilhelmsen KC, Ehlers CL. Associations Between Genomic Variants in Alcohol Dehydrogenase Genes and Alcohol Symptomatology in American Indians and European Americans: Distinctions and Convergence. Alcohol: Clin Exp Res. 2017;41:1695–704.
Peng Q, Bizon C, Gizer IR, Wilhelmsen KC, Ehlers CL. Genetic loci for alcohol-related life events and substance-induced affective symptoms: indexing the “dark side” of addiction. Transl Psychiatry. 2019;9:71.
Peng Q, Wilhelmsen KC, Ehlers CL. Common genetic substrates of alcohol and substance use disorder severity revealed by pleiotropy detection against GWAS catalog in two populations. Addiction Biol. 2020:e12877.
Schuckit MA. The Clinical Implications of Primary Diagnostic Groups Among Alcoholics. Arch Gen Psychiatry. 1985;42:1043–9.
Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI Jr., et al. A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. J Stud Alcohol. 1994;55:149–58.
Schuckit MA, Smith TL, Anthenelli R, Irwin M. Clinical course of alcoholism in 636 male inpatients. Am J Psychiatry. 1993;150:786–92.
Schuckit MA, Smith TL, Danko GP, Reich T, Bucholz KK, Bierut LJ. Similarities in the Clinical Characteristics Related to Alcohol Dependence in Two Populations. Am J Addictions. 2002;11:1–9.
Ehlers CL, Gizer IR, Vieten C, Gilder A, Gilder DA, Stouffer GM, et al. Age at Regular Drinking, Clinical Course, and Heritability of Alcohol Dependence in the San Francisco Family Study: A Gender Analysis. Am J Addictions. 2010;19:101–10.
Ehlers CL, Stouffer GM, Corey L, Gilder DA. The clinical course of DSM-5 alcohol use disorders in young adult native and Mexican Americans. Am J Addictions. 2015;24:713–21.
Schuckit MA, Anthenelli RM, Bucholz KK, Hesselbrock VM, Tipp J. The time course of development of alcohol-related problems in men and women. J Stud Alcohol. 1995;56:218–25.
Venner KL, Miller WR. Progression of alcohol problems in a Navajo sample. J Stud Alcohol. 2001;62:158–65.
Malcolm BP, Hesselbrock MN, Segal B. Multiple Substance Dependence and Course of Alcoholism among Alaska Native Men and Women. Subst Use Misuse 2006;41:729–41.
Scott DM, Williams CD, Cain GE, Kwagyan J, Kalu N, Ehlers CL, et al. Clinical Course of Alcohol Dependence in African Americans. J Addictive Dis. 2008;27:43–50.
Montane-Jaime LK, Shafe S, Joseph R, Moore S, Gilder DA, Crooks H, et al. The Clinical Course of Alcoholism in Trinidad and Tobago. J Stud Alcohol Drugs. 2008;69:834–9.
Bizon C, Spiegel M, Chasse SA, Gizer IR, Li Y, Malc EP, et al. Variant calling in low-coverage whole genome sequencing of a Native American population sample. BMC Genomics. 2014;15:85.
Allison DB, Thiel B, St. Jean P, Elston RC, Infante MC, Schork NJ. Multiple Phenotype Modeling in Gene-Mapping Studies of Quantitative Traits: Power Advantages. Am J Hum Genet. 1998;63:1190–201.
Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. 2012;44:821–4.
Lee S, Emond MJ, Bamshad MJ, Barnes KC, Rieder MJ, Nickerson DA, et al. Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am J Hum Genet. 2012;91:224–37.
Kang HM, Sul JH, Service SK, Zaitlen NA, Kong S-Y, Freimer NB, et al. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42:348–54.
Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89:82–93.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statis Soc B. 1995;57:289–300.
Yekutieli D, Benjamini Y. Resampling-based false discovery rate controlling multiple test procedures for correlated test statistics. J Stat Plan Inference. 1999;82:171–96.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102:15545.
Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JillP, Tamayo P. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst. 2015;1:417–25.
Stouffer S, DeVinney L, Suchmen E. The American soldier: Adjustment during army life. Princeton, US: Princeton University Press; 1949.
LeBlanc M, Zuber V, Thompson WK, Andreassen OA, Frigessi A, Andreassen BK, et al. A correction for sample overlap in genome-wide association studies in a polygenic pleiotropy-informed framework. BMC Genomics. 2018;19:494.
Rentzsch P, Schubach M, Shendure J, Kircher M. CADD-Splice—improving genome-wide variant effect prediction using deep learning-derived splice scores. Genome Med. 2021;13:31.
Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–5.
Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1826.
Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W20.
Ramasamy A, Trabzuni D, Guelfi S, Varghese V, Smith C, Walker R, et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci. 2014;17:1418–28.
Saunders GRB, Wang X, Chen F, Jang S-K, Liu M, Wang C, et al. Genetic diversity fuels gene discovery for tobacco and alcohol use. Nature. 2022;612:720–4.
Okbay A, Wu Y, Wang N, Jayashankar H, Bennett M, Nehzati SM, et al. Polygenic prediction of educational attainment within and between families from genome-wide association analyses in 3 million individuals. Nat Genet. 2022;54:437–49.
Macpherson T, Hikida T. Role of basal ganglia neurocircuitry in the pathology of psychiatric disorders. Psychiatry Clin Neurosci. 2019;73:289–301.
Riblet NB, Gottlieb DJ, Watts BV, Cornelius SL, Fan VS, Shi X, et al. Hypoxia-related risk factors for death by suicide in a national clinical sample. Psychiatry Res. 2019;273:247–51.
Mariani N, Cattane N, Pariante C, Cattaneo A. Gene expression studies in Depression development and treatment: an overview of the underlying molecular mechanisms and biological processes to identify biomarkers. Transl Psychiatry. 2021;11:354.
Young SN. Elevated incidence of suicide in people living at altitude, smokers and patients with chronic obstructive pulmonary disease and asthma: possible role of hypoxia causing decreased serotonin synthesis. J Psychiatry Neurosci. 2013;38:423.
Morales-Medina JC, Witchey SK, Caldwell HK. The Role of Vasopressin in Anxiety and Depression. In: López-Muñoz F, Srinivasan V, de Berardis D, Álamo C, et al., editors. Melatonin, Neuroprotective Agents and Antidepressant Therapy. New Delhi: Springer India; 2016. p. 667–85.
Zhou H, Sealock JM, Sanchez-Roige S, Clarke T-K, Levey DF, Cheng Z, et al. Genome-wide meta-analysis of problematic alcohol use in 435,563 individuals yields insights into biology and relationships with other traits. Nat Neurosci. 2020;23:809–18.
Wang H, Yang J, Schneider JA, De Jager PL, Bennett DA, Zhang H-Y. Genome-wide interaction analysis of pathological hallmarks in Alzheimer’s disease. Neurobiol Aging. 2020;93:61–8.
Lunnon K, Smith R, Hannon E, De Jager PL, Srivastava G, Volta M, et al. Methylomic profiling implicates cortical deregulation of ANK1 in Alzheimer’s disease. Nat Neurosci. 2014;17:1164–70.
De Jager PL, Srivastava G, Lunnon K, Burgess J, Schalkwyk LC, Yu L, et al. Alzheimer’s disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat Neurosci. 2014;17:1156–63.
Liscovitch N, French L. Differential Co-Expression between α-Synuclein and IFN-γ Signaling Genes across Development and in Parkinson’s Disease. PLOS ONE. 2014;9:e115029.
Silverman JS, Skaar JR, Pagano M. SCF ubiquitin ligases in the maintenance of genome stability. Trends Biochem Sci. 2012;37:66–73.
Yang Y, Hadjikyriacou A, Xia Z, Gayatri S, Kim D, Zurita-Lopez C, et al. PRMT9 is a Type II methyltransferase that methylates the splicing factor SAP145. Nat Commun. 2015;6:6428.
Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet. 2019;51:237–44.
Baselmans B, Hammerschlag AR, Noordijk S, Ip H, van der Zee M, de Geus E, et al. The Genetic and Neural Substrates of Externalizing Behavior. Biol Psychiatry Glob Open Sci. 2022;2:389–99.
Karlsson Linnér R, Biroli P, Kong E, Meddens SFW, Wedow R, Fontana MA, et al. Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences. Nat Genet. 2019;51:245–57.
Karlsson Linnér R, Mallard TT, Barr PB, Sanchez-Roige S, Madole JW, Driver MN, et al. Multivariate analysis of 1.5 million people identifies genetic associations with traits related to self-regulation and addiction. Nat Neurosci. 2021;24:1367–76.
Watanabe K, Jansen PR, Savage JE, Nandakumar P, Wang X, Agee M, et al. Genome-wide meta-analysis of insomnia prioritizes genes associated with metabolic and psychiatric pathways. Nat Genet. 2022;54:1125–32.
Blokland GAM, Grove J, Chen C-Y, Cotsapas C, Tobet S, Handa R, et al. Sex-Dependent Shared and Nonshared Genetic Architecture Across Mood and Psychotic Disorders. Biol Psychiatry. 2022;91:102–17.
Aberg KA, Liu Y, Bukszár J, McClay JL, Khachane AN, Andreassen OA, et al. A Comprehensive Family-Based Replication Study of Schizophrenia Genes. JAMA Psychiatry. 2013;70:573–81.
Jansen S, van der Werf IM, Innes AM, Afenjar A, Agrawal PB, Anderson IJ, et al. De novo variants in FBXO11 cause a syndromic form of intellectual disability with behavioral problems and dysmorphisms. Eur J Hum Genet. 2019;27:738–46.
Gregor A, Sadleir LG, Asadollahi R, Azzarello-Burri S, Battaglia A, Ousager LB, et al. De Novo Variants in the F-Box Protein FBXO11 in 20 Individuals with a Variable Neurodevelopmental Disorder. Am J Hum Genet. 2018;103:305–16.
Gouveia C, Gibbons E, Dehghani N, Eapen J, Guerreiro R, Bras J. Genome-wide association of polygenic risk extremes for Alzheimer’s disease in the UK Biobank. Sci Rep. 2022;12:8404.
Dawson DW, Volpert OV, Gillis P, Crawford SE, Xu HJ, Benedict W, et al. Pigment Epithelium-Derived Factor: A Potent Inhibitor of Angiogenesis. Science. 1999;285:245–8.
Crowe S, Wu LE, Economou C, Turpin SM, Matzaris M, Hoehn KL, et al. Pigment Epithelium-Derived Factor Contributes to Insulin Resistance in Obesity. Cell Metab. 2009;10:40–7.
Tian T, Yang Y, Xu B, Qin Y, Zang G, Zhou C, et al. Pigment epithelium-derived factor alleviates depressive-like behaviors in mice by modulating adult hippocampal synaptic growth and Wnt pathway. Prog Neuro-Psychopharmacol Biol Psychiatry. 2020;98:109792.
Bai M, Yu H, Chen C, Xu X, He Y, Wang Y, et al. Pigment epithelium-derived factor may induce antidepressant phenotypes in mice by the prefrontal cortex. Neurosci Lett. 2022;771:136423.
Mann JJ. Neurobiology of suicidal behaviour. Nat Rev Neurosci. 2003;4:819–28.
Arloth J, Eraslan G, Andlauer TFM, Martins J, Iurato S, Kühnel B, et al. DeepWAS: Multivariate genotype-phenotype associations by directly integrating regulatory information using deep learning. PLOS Comput Biol. 2020;16:e1007616.
Gana S, Veggiotti P, Sciacca G, Fedeli C, Bersano A, Micieli G, et al. 19q13.11 cryptic deletion: description of two new cases and indication for a role of WTIP haploinsufficiency in hypospadias. Eur J Hum Genet. 2012;20:852–6.
Tazawa S, Yamato T, Fujikura H, Hiratochi M, Itoh F, Tomae M, et al. SLC5A9/SGLT4, a new Na+-dependent glucose transporter, is an essential transporter for mannose, 1,5-anhydro-D-glucitol, and fructose. Life Sci. 2005;76:1039–50.
Brouwer RM, Klein M, Grasby KL, Schnack HG, Jahanshad N, Teeuw J, et al. Genetic variants associated with longitudinal changes in brain structure across the lifespan. Nat Neurosci. 2022;25:421–32.
Tahir UA, Katz DH, Avila-Pachecho J, Bick AG, Pampana A, Robbins JM, et al. Whole Genome Association Study of the Plasma Metabolome Identifies Metabolites Linked to Cardiometabolic Disease in Black Individuals. Nat Commun. 2022;13:4923.
Argos M, Tong L, Pierce BL, Rakibuz-Zaman M, Ahmed A, Islam T, et al. Genome-wide association study of smoking behaviours among Bangladeshi adults. J Med Genet. 2014;51:327.
van der Meer D, Kaufmann T, Shadrin AA, Makowski C, Frei O, Roelfs D, et al. The genetic architecture of human cortical folding. Sci Adv. 2022;7:eabj9446.
Greenwood TA, Lazzeroni LC, Maihofer AX, Swerdlow NR, Calkins ME, Freedman R, et al. Genome-wide Association of Endophenotypes for Schizophrenia From the Consortium on the Genetics of Schizophrenia (COGS) Study. JAMA Psychiatry. 2019;76:1274–84.
Li QS, Parrado AR, Samtani MN, Narayan VA. Alzheimer’s Disease Neuroimaging I. Variations in the FRA10AC1 Fragile Site and 15q21 Are Associated with Cerebrospinal Fluid Aβ1-42 Level. PLOS ONE. 2015;10:e0134000.
Zhang H, Kranzler HR, Yang BZ, Luo X, Gelernter J. The OPRD1 and OPRK1 loci in alcohol or drug dependence: OPRD1 variation modulates substance dependence risk. Mol Psychiatry. 2008;13:531–43.
Levran O, Londono D, O’Hara K, Nielsen DA, Peles E, Rotrosen J, et al. Genetic susceptibility to heroin addiction: a candidate gene association study. Genes, Brain Behav. 2008;7:720–9.
Lam M, Chen C-Y, Li Z, Martin AR, Bryois J, Ma X, et al. Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat Genet. 2019;51:1670–8.
Kim N, Mickelson JB, Brenner BE, Haws CA, Yurgelun-Todd DA, Renshaw PF. Altitude, Gun Ownership, Rural Areas, and Suicide. Am J Psychiatry. 2011;168:49–54.
Gao G, Li Y, Gee S, Dudley A, Fant J, Crosson C, et al. Down-regulation of Vascular Endothelial Growth Factor and Up-regulation of Pigment Epithelium-derived Factor: A POSSIBLE MECHANISM FOR THE ANTI-ANGIOGENIC ACTIVITY OF PLASMINOGEN KRINGLE 5 *. J Biol Chem. 2002;277:9492–7.
Kang I, Kondo D, Kim J, Lyoo IK, Yurgelun-Todd D, Hwang J, et al. Elevating the level of hypoxia inducible factor may be a new potential target for the treatment of depression. Med Hypotheses. 2021;146:110398.
Bae W-J, Shin M-R, Kang S-K, Zhang J, Kim J-Y, Lee S-C, et al. HIF-2 Inhibition Supresses Inflammatory Responses and Osteoclastic Differentiation in Human Periodontal Ligament Cells. J Cell Biochem. 2015;116:1241–55.
Morris NL, Yeligar SM. Role of HIF-1α in Alcohol-Mediated Multiple Organ Dysfunction. Biomolecules. 2018; 8.
Krishnan HR, Zhang H, Chen Y, Bohnsack JP, Shieh AW, Kusumo H, et al. Unraveling the epigenomic and transcriptomic interplay during alcohol-induced anxiolysis. Mol Psychiatry. 2022;27:4624–32.
Caldwell HK. Neurobiology of Sociability. In: López-Larrea C, editor. Sensing in Nature. New York, NY: Springer US; 2012. p. 187–205.
Kelly AM, Goodson JL. Social functions of individual vasopressin–oxytocin cell groups in vertebrates: What do we really know? Front Neuroendocrinol. 2014;35:512–29.
Oquendo MA, Sullivan GM, Sudol K, Baca-Garcia E, Stanley BH, Sublette ME, et al. Toward a biosignature for suicide. Am J Psychiatry. 2014;171:1259–77.
Mann JJ, Arango VA, Avenevoli S, Brent DA, Champagne FA, Clayton P, et al. Candidate Endophenotypes for Genetic Studies of Suicidal Behavior. Biol Psychiatry. 2009;65:556–63.
Gibbons JL. Electrolytes and Depressive Illness. Postgrad Med J. 1963;39:19.
Ellis GG, Coppen A, Glen AIM. Urine concentration in depressive illness. J Neurol, Neurosurg Amp; Psychiatry. 1971;34:30.
Harper KM, Knapp DJ, Criswell HE, Breese GR. Vasopressin and alcohol: a multifaceted relationship. Psychopharmacology. 2018;235:3363–79.
Zhao F-Q, Keating FA. Functional Properties and Genomics of Glucose Transporters. Curr Genomics. 2007;8:113–28.
Koponen H, Kautiainen H, Leppänen E, Mäntyselkä P, Vanhala M. Association between suicidal behaviour and impaired glucose metabolism in depressive disorders. BMC Psychiatry. 2015;15:163.
Dong R, Haque A, Wu HE, Placide J, Yu L, Zhang X. Sex differences in the association between suicide attempts and glucose disturbances in first-episode and drug naive patients with major depressive disorder. J Affect Disord. 2021;292:559–64.
Gilder DA, Wall TL, Ehlers CL. Comorbidity of select anxiety and affective disorders with alcohol dependence in southwest California Indians. Alcohol Clin Exp Res. 2004;28:1805–13.
Cwik MF, O’Keefe VM, Haroz EE. Suicide in the pediatric population: screening, risk assessment and treatment. Int Rev Psychiatry. 2020;32:254–64.
Funding
This work was supported by the National Institutes of Health (NIH): National Institute on Alcohol Abuse and Alcoholism (NIAAA) R01AA026248 and R01AA027316 to CLE and K25AA025095 to QP, and National Institute on Drug Abuse (NIDA) DP1DA054373 to QP. NIAAA and NIDA had no further role in study design; collection, analysis, and interpretation of data; writing of the report; or the decision to submit the article for publication. The authors report no conflicts of interest, competing interests or undisclosed financial support. The authors alone are responsible for the content and writing of this paper, and the views expressed do not necessarily represent the views of the NIH, NIAAA or NIDA. Open access funding provided by SCELC, Statewide California Electronic Library Consortium.
Author information
Authors and Affiliations
Contributions
QP: Conceptualization, Methodology, Software, Formal analyses, Validation, Writing - original draft, review & editing, Visualization, Funding acquisition. CLE: Data acquisition, Conceptualization, Methodology, Formal analyses, Writing - original draft, review & editing, Supervision, Project administration, Visualization. RAB: Conceptualization, Methodology, Writing-review & editing, DAG: Supervision, Formal analysis, Data curation, Writing-review & editing, KK-J: Conceptualization, Project administration, Supervision, Writing-review & editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The authors assert that all procedures and protocols for the study were carried out in accordance with the latest version of the Declaration of Helsinki. The protocol and procedures were approved by the Institutional Review Board of The Scripps Research Institute and Indian Health Council for the AI tribes participating. Written informed consent was obtained from each participant after the study was fully explained by study staff.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Peng, Q., Gilder, D.A., Bernert, R.A. et al. Genetic factors associated with suicidal behaviors and alcohol use disorders in an American Indian population. Mol Psychiatry (2024). https://doi.org/10.1038/s41380-023-02379-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41380-023-02379-3