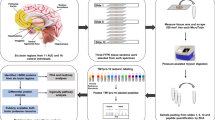
Abstract
Alcohol use disorder (AUD) is a life-threatening disease characterized by compulsive drinking, cognitive deficits, and social impairment that continue despite negative consequences. The inability of individuals with AUD to regulate drinking may involve functional deficits in cortical areas that normally balance actions that have aspects of both reward and risk. Among these, the orbitofrontal cortex (OFC) is critically involved in goal-directed behavior and is thought to maintain a representation of reward value that guides decision making. In the present study, we analyzed post-mortem OFC brain samples collected from age- and sex-matched control subjects and those with AUD using proteomics, bioinformatics, machine learning, and reverse genetics approaches. Of the 4,500+ total unique proteins identified in the proteomics screen, there were 47 proteins that differed significantly by sex that were enriched in processes regulating extracellular matrix and axonal structure. Gene ontology enrichment analysis revealed that proteins differentially expressed in AUD cases were involved in synaptic and mitochondrial function, as well as transmembrane transporter activity. Alcohol-sensitive OFC proteins also mapped to abnormal social behaviors and social interactions. Machine learning analysis of the post-mortem OFC proteome revealed dysregulation of presynaptic (e.g., AP2A1) and mitochondrial proteins that predicted the occurrence and severity of AUD. Using a reverse genetics approach to validate a target protein, we found that prefrontal Ap2a1 expression significantly correlated with voluntary alcohol drinking in male and female genetically diverse mouse strains. Moreover, recombinant inbred strains that inherited the C57BL/6J allele at the Ap2a1 interval consumed higher amounts of alcohol than those that inherited the DBA/2J allele. Together, these findings highlight the impact of excessive alcohol consumption on the human OFC proteome and identify important cross-species cortical mechanisms and proteins that control drinking in individuals with AUD.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Collaborators GBDRF. Global burden of 87 risk factors in 204 countries and territories, 1990-2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet. 2020;396:1223–49.
Spectrum N. Alcohol-related deaths continued to increase in 2021. vol. 15. National Institute of Alcohol Abuse and Alcoholism 2023.
Glantz MD, Bharat C, Degenhardt L, Sampson NA, Scott KM, Lim CCW, et al. The epidemiology of alcohol use disorders cross-nationally: Findings from the World Mental Health Surveys. Addict Behav. 2020;102:106128.
DSM-5. Diagnostic and statistical manual of mental disorders: DSM-5-TR. 5th ed. Text Revision DSM-5-TR edn. American Psychiatric Association: Arlington, VA, 2022.
Koob GF Anhedonia, Hyperkatifeia, and Negative Reinforcement in Substance Use Disorders. Curr Top Behav Neurosci 2022;58:147–65.
Badanich K, Mulholland P, Beckley J, Trantham-Davidson H, Woodward J. Ethanol reduces neuronal excitability of lateral orbitofrontal cortex neurons via a glycine receptor dependent mechanism. Neuropsycopharmacology. 2013;38:1176–88.
Nimitvilai S, Lopez MF, Mulholland PJ, Woodward JJ. Chronic intermittent ethanol exposure enhances the excitability and synaptic plasticity of lateral orbitofrontal cortex neurons and induces a tolerance to the acute inhibitory actions of ethanol. Neuropsychopharmacology. 2016;41:1112–27.
Cannady R, Nimitvilai-Roberts S, Jennings SD, Woodward JJ, Mulholland PJ Distinct Region- and Time-Dependent Functional Cortical Adaptations in C57BL/6J Mice after Short and Prolonged Alcohol Drinking. eNeuro 2020;7.
Moorman DE. The role of the orbitofrontal cortex in alcohol use, abuse, and dependence. Prog Neuropsychopharmacol Biol Psychiatry. 2018;87:85–107.
Shields CN, Gremel CM. Review of orbitofrontal cortex in alcohol dependence: a disrupted cognitive map? Alcohol, Clin Exp Res. 2020;44:1952–64.
Hernandez JS, Moorman DE Orbitofrontal Cortex Encodes Preference for Alcohol. eNeuro 2020;7.
Jokisch D, Roser P, Juckel G, Daum I, Bellebaum C. Impairments in learning by monetary rewards and alcohol-associated rewards in detoxified alcoholic patients. Alcohol, Clin Exp Res. 2014;38:1947–54.
Badanich KA, Fakih ME, Gurina TS, Roy EK, Hoffman JL, Uruena-Agnes AR et al. Reversal learning and experimenter-administered chronic intermittent ethanol exposure in male rats. Psychopharmacology 2016;233:3615–26.
Badanich K, Becker H, Woodward J. Effects of chronic intermittent ethanol exposure on orbitofrontal and medial prefrontal cortex-dependent behaviors in mice. Behav Neurosci. 2011;125:879–91.
McMurray MS, Amodeo LR, Roitman JD. Effects of voluntary alcohol intake on risk preference and behavioral flexibility during rat adolescence. PLoS One. 2014;9:e100697.
Jedema HP, Carter MD, Dugan BP, Gurnsey K, Olsen AS, Bradberry CW. The acute impact of ethanol on cognitive performance in rhesus macaques. Cereb cortex. 2011;21:1783–91.
Renteria R, Baltz ET, Gremel CM. Chronic alcohol exposure disrupts top-down control over basal ganglia action selection to produce habits. Nat Commun. 2018;9:211.
Volkow ND, Wang GJ, Overall JE, Hitzemann R, Fowler JS, Pappas N, et al. Regional brain metabolic response to lorazepam in alcoholics during early and late alcohol detoxification. Alcohol, Clin Exp Res. 1997;21:1278–84.
Myrick H, Anton RF, Li X, Henderson S, Drobes D, Voronin K, et al. Differential brain activity in alcoholics and social drinkers to alcohol cues: relationship to craving. Neuropsychopharmacol: Off Publ Am Coll Neuropsychopharmacol. 2004;29:393–402.
Schacht JP, Yeongbin I, Hoffman M, Voronin KE, Book SW, Anton RF Effects of pharmacological and genetic regulation of COMT activity in alcohol use disorder: a randomized, placebo-controlled trial of tolcapone. Neuropsychopharmacol Offic Publ Am College Neuropsychopharmacol. 2022;47:1953–60.
Gioia DA, Woodward JJ. Altered activity of lateral orbitofrontal cortex neurons in mice following chronic intermittent ethanol exposure. eNeuro 2021;8.
Nimitvilai S, Uys JD, Woodward JJ, Randall PK, Ball LE, Williams RW, et al. Orbitofrontal neuroadaptations and cross-species synaptic biomarkers in heavy-drinking macaques. J Neurosci. 2017;37:3646–60.
Ongur D, Ferry AT, Price JL. Architectonic subdivision of the human orbital and medial prefrontal cortex. J Comp Neurol. 2003;460:425–49.
Li J, Van Vranken JG, Pontano Vaites L, Schweppe DK, Huttlin EL, Etienne C, et al. TMTpro reagents: a set of isobaric labeling mass tags enables simultaneous proteome-wide measurements across 16 samples. Nat methods. 2020;17:399–404.
McAlister GC, Nusinow DP, Jedrychowski MP, Wuhr M, Huttlin EL, Erickson BK, et al. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal Chem. 2014;86:7150–8.
Eng JK, Jahan TA, Hoopmann MR. Comet: an open-source MS/MS sequence database search tool. Proteomics. 2013;13:22–24.
Wilmarth PA, Riviere MA, David LL. Techniques for accurate protein identification in shotgun proteomic studies of human, mouse, bovine, and chicken lenses. J Ocul Biol Dis Info. 2009;2:223–34.
Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S, et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol. 2012;30:918–20.
McDonald WH, Tabb DL, Sadygov RG, MacCoss MJ, Venable J, Graumann J, et al. MS1, MS2, and SQT-three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid Commun Mass Spectrom. 2004;18:2162–8.
Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–92.
Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat methods. 2007;4:207–14.
Plubell DL, Wilmarth PA, Zhao Y, Fenton AM, Minnier J, Reddy AP, et al. Extended Multiplexing of Tandem Mass Tags (TMT) labeling reveals age and high fat diet specific proteome changes in mouse epididymal adipose tissue. Mol Cell Proteom: MCP. 2017;16:873–90.
Perez-Riverol Y, Bai J, Bandla C, Garcia-Seisdedos D, Hewapathirana S, Kamatchinathan S, et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022;50:D543–52.
Le Cao KA, Boitard S, Besse P. Sparse PLS discriminant analysis: biologically relevant feature selection and graphical displays for multiclass problems. BMC Bioinforma. 2011;12:253.
McGuier NS, Rinker JA, Cannady R, Fulmer DB, Jones SR, Hoffman M, et al. Identification and validation of midbrain Kcnq4 regulation of heavy alcohol consumption in rodents. Neuropharmacology. 2018;138:10–19.
Padula AE, Griffin WC 3rd, Lopez MF, Nimitvilai S, Cannady R, McGuier NS, et al. KCNN genes that encode small-conductance Ca2+-Activated K+ channels influence alcohol and drug addiction. Neuropsychopharmacology. 2015;40:1928–39.
Padula AE, Rinker JA, Lopez MF, Mulligan MK, Williams RW, Becker HC, et al. Bioinformatics identification and pharmacological validation of Kcnn3/K(Ca)2 channels as a mediator of negative affective behaviors and excessive alcohol drinking in mice. Transl psychiatry. 2020;10:414.
Rinker JA, Fulmer DB, Trantham-Davidson H, Smith ML, Williams RW, Lopez MF, et al. Differential potassium channel gene regulation in BXD mice reveals novel targets for pharmacogenetic therapies to reduce heavy alcohol drinking. Alcohol. 2017;58:33–45.
McGuier NS, Griffin WC, 3rd, Gass JT, Padula AE, Chesler EJ, Mulholland PJ Kv7 channels in the nucleus accumbens are altered by chronic drinking and are targets for reducing alcohol consumption. Add Biol. 2015;21:1097–112.
Baker EJ, Jay JJ, Bubier JA, Langston MA, Chesler EJ. GeneWeaver: a web-based system for integrative functional genomics. Nucleic acids Res. 2012;40:D1067–76.
Philip VM, Duvvuru S, Gomero B, Ansah TA, Blaha CD, Cook MN, et al. High-throughput behavioral phenotyping in the expanded panel of BXD recombinant inbred strains. Genes Brain Behav. 2010;9:129–59.
Dickson PE, Miller MM, Calton MA, Bubier JA, Cook MN, Goldowitz D, et al. Systems genetics of intravenous cocaine self-administration in the BXD recombinant inbred mouse panel. Psychopharmacology. 2016;233:701–14.
Lopez MF, Miles MF, Williams RW, Becker HC. Variable effects of chronic intermittent ethanol exposure on ethanol drinking in a genetically diverse mouse cohort. Alcohol. 2017;58:73–82.
Faul F, Erdfelder E, Lang AG, Buchner A. G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods. 2007;39:175–91.
Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–3.
Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–11.
Cahill KM, Huo Z, Tseng GC, Logan RW, Seney ML. Improved identification of concordant and discordant gene expression signatures using an updated rank-rank hypergeometric overlap approach. Sci Rep. 2018;8:9588.
Uys JD, McGuier NS, Gass JT, Griffin WC 3rd, Ball LE, Mulholland PJ. Chronic intermittent ethanol exposure and withdrawal leads to adaptations in nucleus accumbens core postsynaptic density proteome and dendritic spines. Addict Biol. 2016;21:560–74.
Kapoor M, Wang JC, Wetherill L, Le N, Bertelsen S, Hinrichs AL, et al. A meta-analysis of two genome-wide association studies to identify novel loci for maximum number of alcoholic drinks. Hum Genet. 2013;132:1141–51.
Grant JD, Agrawal A, Bucholz KK, Madden PA, Pergadia ML, Nelson EC, et al. Alcohol consumption indices of genetic risk for alcohol dependence. Biol psychiatry. 2009;66:795–800.
Saccone NL, Kwon JM, Corbett J, Goate A, Rochberg N, Edenberg HJ, et al. A genome screen of maximum number of drinks as an alcoholism phenotype. Am J Med Genet. 2000;96:632–7.
Schuckit MA, Smith TL, Danko GP, Bucholz KK, Agrawal A, Dick DM, et al. Predictors of subgroups based on maximum drinks per occasion over six years for 833 adolescents and young adults in COGA. J Stud Alcohol Drugs. 2014;75:24–34.
Wang X, Pandey AK, Mulligan MK, Williams EG, Mozhui K, Li Z, et al. Joint mouse-human phenome-wide association to test gene function and disease risk. Nat Commun. 2016;7:10464.
Caruana NJ, Stroud DA. The road to the structure of the mitochondrial respiratory chain supercomplex. Biochem Soc Trans. 2020;48:621–9.
Qin L, Vetreno RP, Crews FT. NADPH oxidase and endoplasmic reticulum stress is associated with neuronal degeneration in orbitofrontal cortex of individuals with alcohol use disorder. Addiction Biol. 2023;28:e13262.
Flatscher-Bader T, van der Brug M, Hwang JW, Gochee PA, Matsumoto I, Niwa S, et al. Alcohol-responsive genes in the frontal cortex and nucleus accumbens of human alcoholics. J Neurochem. 2005;93:359–70.
Sokolov BP, Jiang L, Trivedi NS, Aston C. Transcription profiling reveals mitochondrial, ubiquitin and signaling systems abnormalities in postmortem brains from subjects with a history of alcohol abuse or dependence. J Neurosci Res. 2003;72:756–67.
Liu J, Lewohl JM, Dodd PR, Randall PK, Harris RA, Mayfield RD. Gene expression profiling of individual cases reveals consistent transcriptional changes in alcoholic human brain. J Neurochem. 2004;90:1050–8.
Shang P, Lindberg D, Starski P, Peyton L, Hong SI, Choi S, et al. Chronic alcohol exposure induces aberrant mitochondrial morphology and inhibits respiratory capacity in the medial prefrontal cortex of mice. Front Neurosci. 2020;14:561173.
Jung ME, Metzger DB. Aberrant histone acetylation promotes mitochondrial respiratory suppression in the brain of alcoholic rats. J Pharm Exp Ther. 2015;352:258–66.
Reddy VD, Padmavathi P, Kavitha G, Saradamma B, Varadacharyulu N. Alcohol-induced oxidative/nitrosative stress alters brain mitochondrial membrane properties. Mol Cell Biochem. 2013;375:39–47.
Vetreno RP, Qin L, Coleman LG Jr., Crews FT. Increased Toll-like Receptor-MyD88-NFkappaB-Proinflammatory neuroimmune signaling in the orbitofrontal cortex of humans with alcohol use disorder. Alcohol, Clin Exp Res. 2021;45:1747–61.
Miguel-Hidalgo JJ, Overholser JC, Meltzer HY, Stockmeier CA, Rajkowska G. Reduced glial and neuronal packing density in the orbitofrontal cortex in alcohol dependence and its relationship with suicide and duration of alcohol dependence. Alcohol, Clin Exp Res. 2006;30:1845–55.
Ruggiero A, Katsenelson M, Slutsky I. Mitochondria: new players in homeostatic regulation of firing rate set points. Trends Neurosci. 2021;44:605–18.
Styr B, Gonen N, Zarhin D, Ruggiero A, Atsmon R, Gazit N, et al. Mitochondrial regulation of the hippocampal firing rate set point and seizure susceptibility. Neuron. 2019;102:1009–24.e1008.
Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD. Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J Neurosci. 2012;32:1884–97.
Wolen AR, Phillips CA, Langston MA, Putman AH, Vorster PJ, Bruce NA, et al. Genetic dissection of acute ethanol responsive gene networks in prefrontal cortex: functional and mechanistic implications. PloS one. 2012;7:e33575.
Lovinger DM. Presynaptic ethanol actions: potential roles in ethanol seeking. Handb Exp Pharm. 2018;248:29–54.
Le Berre AP. Emotional processing and social cognition in alcohol use disorder. Neuropsychology. 2019;33:808–21.
Willis ML, Palermo R, Burke D, McGrillen K, Miller L. Orbitofrontal cortex lesions result in abnormal social judgements to emotional faces. Neuropsychologia. 2010;48:2182–7.
Watson KK, Platt ML. Social signals in primate orbitofrontal cortex. Curr Biol. 2012;22:2268–73.
Azzi JC, Sirigu A, Duhamel JR. Modulation of value representation by social context in the primate orbitofrontal cortex. Proc Natl Acad Sci USA. 2012;109:2126–31.
Unger EK, Keller JP, Altermatt M, Liang R, Matsui A, Dong C, et al. Directed evolution of a selective and sensitive serotonin sensor via machine learning. Cell. 2020;183:1986–2002.e1926.
Kuniishi H, Nakatake Y, Sekiguchi M, Yamada M. Adolescent social isolation induces distinct changes in the medial and lateral OFC-BLA synapse and social and emotional alterations in adult mice. Neuropsychopharmacol: Off Publ Am Coll Neuropsychopharmacol. 2022;47:1597–607.
Jennings JH, Kim CK, Marshel JH, Raffiee M, Ye L, Quirin S, et al. Interacting neural ensembles in orbitofrontal cortex for social and feeding behaviour. Nature. 2019;565:645–9.
Gautier M, Pabst A, Maurage P. Social decision making in severe alcohol use disorder: Scoping review and experimental perspectives. Alcohol, Clin Exp Res. 2021;45:1548–59.
Charlet K, Schlagenhauf F, Richter A, Naundorf K, Dornhof L, Weinfurtner CE, et al. Neural activation during processing of aversive faces predicts treatment outcome in alcoholism. Addiction Biol. 2014;19:439–51.
Bora E, Zorlu N. Social cognition in alcohol use disorder: a meta-analysis. Addiction. 2017;112:40–8.
Valmas MM, Mosher Ruiz S, Gansler DA, Sawyer KS, Oscar-Berman M. Social cognition deficits and associations with drinking history in alcoholic men and women. Alcohol, Clin Exp Res. 2014;38:2998–3007.
Schmidt T, Roser P, Ze O, Juckel G, Suchan B, Thoma P. Cortical thickness and trait empathy in patients and people at high risk for alcohol use disorders. Psychopharmacology. 2017;234:3521–33.
Hulvershorn LA, Finn P, Hummer TA, Leibenluft E, Ball B, Gichina V, et al. Cortical activation deficits during facial emotion processing in youth at high risk for the development of substance use disorders. Drug Alcohol Depend. 2013;131:230–7.
Hill SY, Kostelnik B, Holmes B, Goradia D, McDermott M, Diwadkar V, et al. fMRI BOLD response to the eyes task in offspring from multiplex alcohol dependence families. Alcohol, Clin Exp Res. 2007;31:2028–35.
Hill SY, Wellman JL, Zezza N, Steinhauer SR, Sharma V, Holmes B Epigenetic Effects in HPA Axis Genes Associated with Cortical Thickness, ERP Components and SUD Outcome. Behav Sci (Basel) 2022;12:347.
Hill SY, Wang S, Kostelnik B, Carter H, Holmes B, McDermott M, et al. Disruption of orbitofrontal cortex laterality in offspring from multiplex alcohol dependence families. Biol Psychiatry. 2009;65:129–36.
Xin J, Zhang Y, Tang Y, Yang Y. Brain differences between men and women: evidence from deep learning. Front Neurosci. 2019;13:185.
Ritchie SJ, Cox SR, Shen X, Lombardo MV, Reus LM, Alloza C, et al. Sex differences in the adult human brain: evidence from 5216 UK biobank participants. Cereb Cortex. 2018;28:2959–75.
Batzdorf CS, Morr AS, Bertalan G, Sack I, Silva RV, Infante-Duarte C Sexual Dimorphism in Extracellular Matrix Composition and Viscoelasticity of the Healthy and Inflamed Mouse Brain. Biology (Basel) 2022;11:230.
Nazlee N, Waiter GD, Sandu AL. Age-associated sex and asymmetry differentiation in hemispheric and lobar cortical ribbon complexity across adulthood: A UK Biobank imaging study. Hum Brain Mapp. 2022;44:49–65.
Heilbronner SR, Haber SN. Frontal cortical and subcortical projections provide a basis for segmenting the cingulum bundle: implications for neuroimaging and psychiatric disorders. J Neurosci: Off J Soc Neurosci. 2014;34:10041–54.
Guo J, Bertalan G, Meierhofer D, Klein C, Schreyer S, Steiner B, et al. Brain maturation is associated with increasing tissue stiffness and decreasing tissue fluidity. Acta Biomater. 2019;99:433–42.
Arani A, Murphy MC, Glaser KJ, Manduca A, Lake DS, Kruse SA, et al. Measuring the effects of aging and sex on regional brain stiffness with MR elastography in healthy older adults. Neuroimage. 2015;111:59–64.
Sack I, Beierbach B, Wuerfel J, Klatt D, Hamhaber U, Papazoglou S, et al. The impact of aging and gender on brain viscoelasticity. Neuroimage. 2009;46:652–7.
Wuerfel J, Paul F, Beierbach B, Hamhaber U, Klatt D, Papazoglou S, et al. MR-elastography reveals degradation of tissue integrity in multiple sclerosis. Neuroimage. 2010;49:2520–5.
Mulholland PJ, Chandler LJ, Kalivas PW. Signals from the fourth dimension regulate drug relapse. Trends Neurosci. 2016;39:472–85.
Lasek AW. Effects of ethanol on brain extracellular matrix: implications for alcohol use disorder. Alcohol, Clin Exp Res. 2016;40:2030–42.
Kruyer A, Chioma VC, Kalivas PW. The opioid-addicted tetrapartite synapse. Biol Psychiatry. 2020;87:34–43.
Seney ML, Kim SM, Glausier JR, Hildebrand MA, Xue X, Zong W, et al. Transcriptional alterations in dorsolateral prefrontal cortex and nucleus accumbens implicate neuroinflammation and synaptic remodeling in opioid use disorder. Biol Psychiatry. 2021;90:550–62.
Sheedy D, Garrick T, Dedova I, Hunt C, Miller R, Sundqvist N, et al. An Australian Brain Bank: a critical investment with a high return! Cell Tissue Bank. 2008;9:205–16.
Acknowledgements
Tissues were received from the New South Wales Brain Tissue Resource Centre at the University of Sydney which is supported by the University of Sydney. Research reported in this publication was supported by the National Institute of Alcohol Abuse and Alcoholism of the National Institutes of Health under Award Number R28AA012725. The content is solely the responsibility of the authors and does not represent the official views of the National Institutes of Health. NIH grants to PJM (R01 AA023288) and Charleston Alcohol Research Center to PJM and JJW (P50 AA010761) supported this work. Data were acquired by LEB (MUSC Mass Spectrometry Facility) with support from NIH grants (S10 OD025126 and P30GM140964). Proteomic data analysis was performed by PW (OHSU Proteomics Shared Resource) with partial support from NIH core grants P30EY010572 and P30CA069533.
Author information
Authors and Affiliations
Contributions
PJM and JJW conceived and coordinated the project. LEB designed and conducted the TMTpro experiment. PAW processed the proteomics data. SB and PJM analyzed the data. PJM performed the concordance analysis and bioinformatics and reverse genetics experiments. CM performed the machine learning analysis. PJM wrote the manuscript. PJM, JJW, LEB, SB, PAW, and CM revised and edited the manuscript. PJM supervised the overall project and provided funding and resources.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mulholland, P.J., Berto, S., Wilmarth, P.A. et al. Adaptor protein complex 2 in the orbitofrontal cortex predicts alcohol use disorder. Mol Psychiatry 28, 4766–4776 (2023). https://doi.org/10.1038/s41380-023-02236-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-023-02236-3