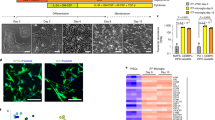
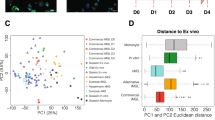
Abstract
Chemogenetic approaches using Designer Receptors Exclusively Activated by Designer Drugs (DREADD, a family of engineered GPCRs) were recently employed in microglia. Here, we used Cx3cr1CreER/+:R26hM4Di/+ mice to express Gi-DREADD (hM4Di) on CX3CR1+ cells, comprising microglia and some peripheral immune cells, and found that activation of hM4Di on long-lived CX3CR1+ cells induced hypolocomotion. Unexpectedly, Gi-DREADD-induced hypolocomotion was preserved when microglia were depleted. Consistently, specific activation of microglial hM4Di cannot induce hypolocomotion in Tmem119CreER/+:R26hM4Di/+ mice. Flow cytometric and histological analysis showed hM4Di expression in peripheral immune cells, which may be responsible for the hypolocomotion. Nevertheless, depletion of splenic macrophages, hepatic macrophages, or CD4+ T cells did not affect Gi-DREADD-induced hypolocomotion. Our study demonstrates that rigorous data analysis and interpretation are needed when using Cx3cr1CreER/+ mouse line to manipulate microglia.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Cserep C, Posfai B, Denes A. Shaping neuronal fate: functional heterogeneity of direct microglia-neuron interactions. Neuron. 2021;109:222–40.
Wolf SA, Boddeke HW, Kettenmann H. Microglia in physiology and disease. Annu Rev Physiol. 2017;79:619–43.
Eyo UB, Wu LJ. Bidirectional microglia-neuron communication in the healthy brain. Neural Plast. 2013;2013:456857.
Umpierre AD, Wu LJ. How microglia sense and regulate neuronal activity. Glia. 2021;69:1637–53.
Haque ME, Kim IS, Jakaria M, Akther M, Choi DK. Importance of GPCR-mediated microglial activation in Alzheimer’s disease. Front Cell Neurosci. 2018;12:258.
Hsiao CC, Sankowski R, Prinz M, Smolders J, Huitinga I, Hamann J. GPCRomics of homeostatic and disease-associated human microglia. Front Immunol. 2021;12:674189.
Eyo UB, Wu LJ. Microglia: lifelong patrolling immune cells of the brain. Prog Neurobiol. 2019;179:101614.
Merlini M, Rafalski VA, Ma K, Kim KY, Bushong EA, Rios Coronado PE, et al. Microglial Gi-dependent dynamics regulate brain network hyperexcitability. Nat Neurosci. 2021;24:19–23.
Armbruster BN, Li X, Pausch MH, Herlitze S, Roth BL. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc Natl Acad Sci USA. 2007;104:5163–8.
Roth BL. DREADDs for neuroscientists. Neuron. 2016;89:683–94.
Grace PM, Strand KA, Galer EL, Urban DJ, Wang X, Baratta MV, et al. Morphine paradoxically prolongs neuropathic pain in rats by amplifying spinal NLRP3 inflammasome activation. Proc Natl Acad Sci USA. 2016;113:E3441–50.
Klawonn AM, Fritz M, Castany S, Pignatelli M, Canal C, Simila F, et al. Microglial activation elicits a negative affective state through prostaglandin-mediated modulation of striatal neurons. Immunity. 2021;54:225–34.
Saika F, Matsuzaki S, Kishioka S, Kiguchi N. Chemogenetic activation of CX3CR1-expressing spinal microglia using Gq-DREADD elicits mechanical allodynia in male mice. Cells. 2021;10:874.
Parusel S, Yi MH, Hunt CL, Wu LJ. Chemogenetic and optogenetic manipulations of microglia in chronic pain. Neurosci Bull. 2023;39:368–78
Yi MH, Liu YU, Liu K, Chen T, Bosco DB, Zheng J, et al. Chemogenetic manipulation of microglia inhibits neuroinflammation and neuropathic pain in mice. Brain Behav Immun. 2021;92:78–89.
Chen T, Lennon VA, Liu YU, Bosco DB, Li Y, Yi MH, et al. Astrocyte-microglia interaction drives evolving neuromyelitis optica lesion. J Clin Invest. 2020;130:4025–38.
Haynes SE, Hollopeter G, Yang G, Kurpius D, Dailey ME, Gan WB, et al. The P2Y12 receptor regulates microglial activation by extracellular nucleotides. Nat Neurosci. 2006;9:1512–9.
Peng J, Liu Y, Umpierre AD, Xie M, Tian DS, Richardson JR, et al. Microglial P2Y12 receptor regulates ventral hippocampal CA1 neuronal excitability and innate fear in mice. Mol Brain. 2019;12:71.
Lowery RL, Mendes MS, Sanders BT, Murphy AJ, Whitelaw BS, Lamantia CE, et al. Loss of P2Y12 has behavioral effects in the adult mouse. Int J Mol Sci. 2021;22:1868.
Schubert I, Ahlbrand R, Winter A, Vollmer L, Lewkowich I, Sah R. Enhanced fear and altered neuronal activation in forebrain limbic regions of CX3CR1-deficient mice. Brain Behav Immun. 2018;68:34–43.
Pozo-Rodrigalvarez A, Ollaranta R, Skoog J, Pekny M, Pekna M. Hyperactive behavior and altered brain morphology in adult complement C3a receptor deficient mice. Front Immunol. 2021;12:604812.
Parkhurst CN, Yang G, Ninan I, Savas JN, Yates JR 3rd, Lafaille JJ, et al. Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell. 2013;155:1596–609.
Goldmann T, Wieghofer P, Muller PF, Wolf Y, Varol D, Yona S, et al. A new type of microglia gene targeting shows TAK1 to be pivotal in CNS autoimmune inflammation. Nat Neurosci. 2013;16:1618–26.
Yona S, Kim KW, Wolf Y, Mildner A, Varol D, Breker M, et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38:79–91.
Elmore MR, Najafi AR, Koike MA, Dagher NN, Spangenberg EE, Rice RA, et al. Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron. 2014;82:380–97.
Lei F, Cui N, Zhou C, Chodosh J, Vavvas DG, Paschalis EI. CSF1R inhibition by a small-molecule inhibitor is not microglia specific; affecting hematopoiesis and the function of macrophages. Proc Natl Acad Sci USA. 2020;117:23336–8.
Kaiser T, Feng G. Tmem119-EGFP and Tmem119-CreERT2 transgenic mice for labeling and manipulating microglia. eNeuro. 2019;6:ENEURO.0448-18.
Bennett ML, Bennett FC, Liddelow SA, Ajami B, Zamanian JL, Fernhoff NB, et al. New tools for studying microglia in the mouse and human CNS. Proc Natl Acad Sci USA. 2016;113:E1738–46.
Zhan L, Fan L, Kodama L, Sohn PD, Wong MY, Mousa GA, et al. A MAC2-positive progenitor-like microglial population is resistant to CSF1R inhibition in adult mouse brain. Elife. 2020;9:e51796.
Tabula Muris C, Overall C, Logistical C, Organ C, processing, Library p. et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018;562:367–72.
Bosco DB, Tian DS, Wu LJ. Neuroimmune interaction in seizures and epilepsy: focusing on monocyte infiltration. FEBS J. 2020;287:4822–37.
Shichita T, Ooboshi H, Yoshimura A. Neuroimmune mechanisms and therapies mediating post-ischaemic brain injury and repair. Nat Rev Neurosci. 2023;24:299–312.
Shi Z, Yu P, Lin WJ, Chen S, Hu X, Chen S, et al. Microglia drive transient insult-induced brain injury by chemotactic recruitment of CD8(+) T lymphocytes. Neuron. 2023;111:696–710.
Saika F, Matsuzaki S, Kobayashi D, Ideguchi Y, Nakamura TY, Kishioka S, et al. Chemogenetic regulation of CX3CR1-expressing microglia using Gi-DREADD exerts sex-dependent anti-allodynic effects in mouse models of neuropathic pain. Front Pharmacol. 2020;11:925.
Biewenga J, van der Ende MB, Krist LF, Borst A, Ghufron M, van Rooijen N. Macrophage depletion in the rat after intraperitoneal administration of liposome-encapsulated clodronate: depletion kinetics and accelerated repopulation of peritoneal and omental macrophages by administration of Freund’s adjuvant. Cell Tissue Res. 1995;280:189–96.
Arnold L, Perrin H, de Chanville CB, Saclier M, Hermand P, Poupel L, et al. CX3CR1 deficiency promotes muscle repair and regeneration by enhancing macrophage ApoE production. Nat Commun. 2015;6:8972.
Zhao W, Lu H, Wang X, Ransohoff RM, Zhou L. CX3CR1 deficiency delays acute skeletal muscle injury repair by impairing macrophage functions. FASEB J. 2016;30:380–93.
Ilanges A, Shiao R, Shaked J, Luo JD, Yu X, Friedman JM. Brainstem ADCYAP1(+) neurons control multiple aspects of sickness behaviour. Nature. 2022;609:761–71.
Sun L, Ye RD. Role of G protein-coupled receptors in inflammation. Acta Pharmacol Sin. 2012;33:342–50.
Elmquist JK, Scammell TE, Saper CB. Mechanisms of CNS response to systemic immune challenge: the febrile response. Trends Neurosci. 1997;20:565–70.
Wettschureck N, Offermanns S. Mammalian G proteins and their cell type specific functions. Physiol Rev. 2005;85:1159–204.
Grace PM, Wang X, Strand KA, Baratta MV, Zhang Y, Galer EL, et al. DREADDed microglia in pain: implications for spinal inflammatory signaling in male rats. Exp Neurol. 2018;304:125–31.
Bernier LP, Bohlen CJ, York EM, Choi HB, Kamyabi A, Dissing-Olesen L, et al. Nanoscale surveillance of the brain by microglia via cAMP-regulated filopodia. Cell Rep. 2019;27:2895–908.
Binning W, Hogan-Cann AE, Yae Sakae D, Maksoud M, Ostapchenko V, Al-Onaizi M, et al. Chronic hM3Dq signaling in microglia ameliorates neuroinflammation in male mice. Brain Behav Immun. 2020;88:791–801.
Wendeln AC, Degenhardt K, Kaurani L, Gertig M, Ulas T, Jain G, et al. Innate immune memory in the brain shapes neurological disease hallmarks. Nature. 2018;556:332–8.
Van Hove H, Antunes ARP, De Vlaminck K, Scheyltjens I, Van Ginderachter JA, Movahedi K. Identifying the variables that drive tamoxifen-independent CreERT2 recombination: implications for microglial fate mapping and gene deletions. Eur J Immunol. 2020;50:459–63.
Chappell-Maor L, Kolesnikov M, Kim JS, Shemer A, Haimon Z, Grozovski J, et al. Comparative analysis of CreER transgenic mice for the study of brain macrophages: a case study. Eur J Immunol. 2020;50:353–62.
Van Hove H, Martens L, Scheyltjens I, De Vlaminck K, Pombo Antunes AR, De Prijck S, et al. A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nat Neurosci. 2019;22:1021–35.
Kim JS, Kolesnikov M, Peled-Hajaj S, Scheyltjens I, Xia Y, Trzebanski S, et al. A binary cre transgenic approach dissects microglia and CNS border-associated macrophages. Immunity. 2021;54:176–90.
Masuda T, Amann L, Sankowski R, Staszewski O, Lenz M, DE P, et al. Novel Hexb-based tools for studying microglia in the CNS. Nat Immunol. 2020;21:802–15.
McKinsey GL, Lizama CO, Keown-Lang AE, Niu A, Santander N, Larpthaveesarp A, et al. A new genetic strategy for targeting microglia in development and disease. Elife. 2020;9:e54590.
Davalos D, Grutzendler J, Yang G, Kim JV, Zuo Y, Jung S, et al. ATP mediates rapid microglial response to local brain injury in vivo. Nat Neurosci. 2005;8:752–8.
Van Rooijen N, Sanders A. Liposome mediated depletion of macrophages: mechanism of action, preparation of liposomes and applications. J Immunol Methods. 1994;174:83–93.
Hao Y, Hao S, Andersen-Nissen E, Mauck WM 3rd, Zheng S, Butler A, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–87.
Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16:1289–96.
Acknowledgements
We thank Dr. Vanda A. Lennon (Mayo Clinic), Dr. Doo-Sup Choi (Mayo Clinic) and Dr. Bryan Roth (University of North Carolina) for thoughtful comments. We also thank members of the Wu lab for insightful discussions.
Funding
This work was supported by the following grants from the National Institutes of Health: R35NS132326 (L-JW), R01ES033892 (JRR, L-JW), and K99NS126417 (ADU).
Author information
Authors and Affiliations
Contributions
SZ, JZ, and L-JW designed experiments and wrote the manuscript. SZ and JZ performed most of the experiments. LW performed data mining and analyzed RNA sequencing data. ADU, SP, and MX performed some experiments. SZ and AD analyzed data. KA, AJJ, and JRR provided resources for specific experiments.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhao, S., Zheng, J., Wang, L. et al. Chemogenetic manipulation of CX3CR1+ cells transiently induces hypolocomotion independent of microglia. Mol Psychiatry 28, 2857–2871 (2023). https://doi.org/10.1038/s41380-023-02128-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-023-02128-6