Abtract
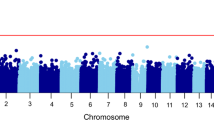
Studies conducted in psychotic disorders have shown that DNA-methylation (DNAm) is sensitive to the impact of Childhood Adversity (CA). However, whether it mediates the association between CA and psychosis is yet to be explored. Epigenome wide association studies (EWAS) using the Illumina Infinium-Methylation EPIC array in peripheral blood tissue from 366 First-episode of psychosis and 517 healthy controls was performed. Adversity scores were created for abuse, neglect and composite adversity with the Childhood Trauma Questionnaire (CTQ). Regressions examining (I) CTQ scores with psychosis; (II) with DNAm EWAS level and (III) between DNAm and caseness, adjusted for a variety of confounders were conducted. Divide-Aggregate Composite-null Test for the composite null-hypothesis of no mediation effect was conducted. Enrichment analyses were conducted with missMethyl package and the KEGG database. Our results show that CA was associated with psychosis (Composite: OR = 1.68; p = <0.001; abuse: OR = 2.16; p < 0.001; neglect: OR = 2.27; p = <0.001). None of the CpG sites significantly mediated the adversity-psychosis association after Bonferroni correction (p < 8.1 × 10−8). However, 28, 34 and 29 differentially methylated probes associated with 21, 27, 20 genes passed a less stringent discovery threshold (p < 5 × 10−5) for composite, abuse and neglect respectively, with a lack of overlap between abuse and neglect. These included genes previously associated to psychosis in EWAS studies, such as PANK1, SPEG TBKBP1, TSNARE1 or H2R. Downstream gene ontology analyses did not reveal any biological pathways that survived false discovery rate correction. Although at a non-significant level, DNAm changes in genes previously associated with schizophrenia in EWAS studies may mediate the CA-psychosis association. These results and associated involved processes such as mitochondrial or histaminergic disfunction, immunity or neural signalling requires replication in well powered samples. The lack of overlap between mediating genes associated with abuse and neglect suggests differential biological trajectories linking CA subtypes and psychosis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Varese F, Smeets F, Drukker M, Lieverse R, Lataster T, Viechtbauer W, et al. Childhood adversities increase the risk of psychosis: a meta-analysis of patient-control, prospective- and cross-sectional cohort studies. Schizophr Bull. 2012;38:661–71.
Rodriguez V, Aas M, Vorontsova N, Trotta G, Gadelrab R, Rooprai NK, et al. Exploring the Interplay Between Adversity, Neurocognition, Social Cognition, and Functional Outcome in People With Psychosis: A Narrative Review. Front Psychiatry. 2021;12:596949.
Christy A, Cavero D, Navajeeva S, Murray-O’Shea R, Rodriguez V, Aas M, et al. Association Between Childhood Adversity and Functional Outcomes in People With Psychosis: A Meta-analysis. Schizophr Bull. 2023;49:285–96.
Alameda L, Christy A, Rodriguez V, Salazar de Pablo G, Thrush M, Shen Y, et al. Association Between Specific Childhood Adversities and Symptom Dimensions in People With Psychosis: Systematic Review and Meta-Analysis. Schizophr Bull. 2021;47:975–85.
Aas M, Haukvik UK, Djurovic S, Tesli M, Athanasiu L, Bjella T, et al. Interplay between childhood trauma and BDNF val66met variants on blood BDNF mRNA levels and on hippocampus subfields volumes in schizophrenia spectrum and bipolar disorders. J Psychiatr Res. 2014;59:14–21.
Howes OD, McCutcheon R, Owen MJ, Murray RM. The Role of Genes, Stress, and Dopamine in the Development of Schizophrenia. Biol Psychiatry. 2017;81:9–20.
Ruby E, Polito S, McMahon K, Gorovitz M, Corcoran C, Malaspina D. Pathways Associating Childhood Trauma to the Neurobiology of Schizophrenia. Front Psychol Behav Sci. 2014;3:1–17.
Alameda L, Fournier M, Khadimallah I, Griffa A, Cleusix M, Jenni R, et al. Redox dysregulation as a link between childhood trauma and psychopathological and neurocognitive profile in patients with early psychosis. Proc Natl Acad Sci. 2018;115:12495–12500.
Alameda L, Rodriguez V, Carr E, Aas M, Trotta G, Marino P, et al. A systematic review on mediators between adversity and psychosis: potential targets for treatment. Psychol Med. 2020;50:1966–76.
Binder EB. Dissecting the molecular mechanisms of gene x environment interactions: implications for diagnosis and treatment of stress-related psychiatric disorders. Eur J Psychotraumatol. 2017;8:1412745.
Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33:245–54.
Alameda L, Trotta G, Quigley H, Rodriguez V, Gadelrab R, Dwir D, et al. Can epigenetics shine a light on the biological pathways underlying major mental disorders? Psychol Med. 2022;52:1645–65.
Baron RM, Kenny DA. The moderator–mediator variable distinction in social psychological research: Conceptual, strategic, and statistical considerations. J Personal Soc Psychol. 1986;51:1173.
Sobel ME. Asymptotic confidence intervals for indirect effects in structural equation models. Sociol Methodol. 1982;13:290–312.
Barfield R, Shen J, Just AC, Vokonas PS, Schwartz J, Baccarelli AA, et al. Testing for the indirect effect under the null for genome‐wide mediation analyses. Genet Epidemiol. 2017;41:824–33.
Liu Z, Shen J, Barfield R, Schwartz J, Baccarelli AA, Lin X. Large-Scale Hypothesis Testing for Causal Mediation Effects with Applications in Genome-wide Epigenetic Studies. J Am Stat Assoc. 2022;117:67–81.
Liu Z, Shen J, Barfield R, Schwartz J, Baccarelli AA, Lin X. Large-scale hypothesis testing for causal mediation effects with applications in genome-wide epigenetic studies. J Am Stat Assoc. 2022;117:67–81.
Gayer-Anderson C, Jongsma HE, Di Forti M, Quattrone D, Velthorst E, de Haan L, et al. The EUropean Network of National Schizophrenia Networks Studying Gene-Environment Interactions (EU-GEI): Incidence and First-Episode Case-Control Programme. Soc Psychiatry Psychiatr Epidemiol. 2020;55:645–57.
Quattrone D, Di Forti M, Gayer-Anderson C, Ferraro L, Jongsma HE, Tripoli G, et al. Transdiagnostic dimensions of psychopathology at first episode psychosis: findings from the multinational EU-GEI study. Psychological Med. 2018;1–14.
McGuffin P, Farmer A, Harvey I. A polydiagnostic application of operational criteria in studies of psychotic illness: development and reliability of the OPCRIT system. Arch Gen Psychiatry. 1991;48:764–70.
Mallett R, Leff J, Bhugra D, Pang D, Zhao JH. Social environment, ethnicity and schizophrenia. A case-control study. Soc psychiatry Psychiatr Epidemiol. 2002;37:329–35.
Bernstein DP, Stein JA, Newcomb MD, Walker E, Pogge D, Ahluvalia T, et al. Development and validation of a brief screening version of the Childhood Trauma Questionnaire. Child Abus Negl. 2003;27:169–90.
Bernstein DP, Fink L, Handelsman L, Foote J, Lovejoy M, Wenzel K, et al. Initial reliability and validity of a new retrospective measure of child abuse and neglect. Am J Psychiatry. 1994;151:1132–6.
Aas M, Alameda L, Di Forti M, Quattrone D, Dazzan P, Trotta A, et al. Synergistic effects of childhood adversity and polygenic risk in first-episode psychosis: the EU-GEI study. Psychological Med. 2021;1–9. https://doi.org/10.1017/S0033291721003664.
Perroud N, Paoloni-Giacobino A, Prada P, Olie E, Salzmann A, Nicastro R. Increased methylation of glucocorticoid receptor gene (NR3C1) in adults with a history of childhood maltreatment: a link with the severity and type of trauma. Transl Psychiatry, 2011;1:e59.
Marzi SJ, Sugden K, Arseneault L, Belsky DW, Burrage J, Corcoran DL, et al. Analysis of DNA Methylation in Young People: Limited Evidence for an Association Between Victimization Stress and Epigenetic Variation in Blood. Am J Psychiatry. 2018. appiajp201717060693.
Bowtell DD. Rapid isolation of eukaryotic DNA. Anal Biochem. 1987;162:463–5.
Jeanpierre M. A rapid method for the purification of DNA from blood. Nucleic Acids Res. 1987;15:9611.
Pidsley R, Viana J, Hannon E, Spiers H, Troakes C, Al-Saraj S, et al. Methylomic profiling of human brain tissue supports a neurodevelopmental origin for schizophrenia. Genome Biol. 2014;15:483.
Bhering LL. Rbio: A tool for biometric and statistical analysis using the R platform. Crop Breed Appl Biotechnol. 2017;17:187–90.
Pidsley R, Wong CC, Volta M, Lunnon K, Mill J, Schalkwyk LC. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics. 2013;14:293.
Hannon E, Dempster EL, Mansell G, Burrage J, Bass N, Bohlken MM, et al. DNA methylation meta-analysis reveals cellular alterations in psychosis and markers of treatment-resistant schizophrenia. eLife. 2021;10:e58430.
Turner SD. qqman: an R package for visualizing GWAS results using QQ and manhattan plots. Biorxiv. 2014. https://doi.org/10.1101/005165.
Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinforma. 2012;13:1–16.
Elliott HR, Tillin T, McArdle WL, Ho K, Duggirala A, Frayling TM, et al. Differences in smoking associated DNA methylation patterns in South Asians and Europeans. Clin Epigenetics. 2014;6:1–10.
Dietz PM, Homa D, England LJ, Burley K, Tong VT, Dube SR, et al. Estimates of nondisclosure of cigarette smoking among pregnant and nonpregnant women of reproductive age in the United States. Am J Epidemiol. 2011;173:355–9.
Spencer K, Cowans NJ. Accuracy of self‐reported smoking status in first trimester aneuploidy screening. Prenat Diagnosis. 2013;33:245–50.
Kandaswamy R, Hannon E, Arseneault L, Mansell G, Sugden K, Williams B, et al. DNA methylation signatures of adolescent victimization: analysis of a longitudinal monozygotic twin sample. Epigenetics 2021;16:1169–86.
Sammallahti S, Cortes Hidalgo AP, Tuominen S, Malmberg A, Mulder RH, Brunst KJ, et al. Maternal anxiety during pregnancy and newborn epigenome-wide DNA methylation. Mol Psychiatry. 2021;26:1832–45.
Phipson B, Maksimovic J, Oshlack A. missMethyl: an R package for analyzing data from Illumina’s HumanMethylation450 platform. Bioinformatics. 2016;32:286–8.
Zou D, Qiu Y, Li R, Meng Y, Wu Y. A Novel Schizophrenia Diagnostic Model Based on Statistically Significant Changes in Gene Methylation in Specific Brain Regions. Biomed Res Int. 2020;2020:8047146.
Li M, Li Y, Qin H, Tubbs JD, Li M, Qiao C, et al. Genome-wide DNA methylation analysis of peripheral blood cells derived from patients with first-episode schizophrenia in the Chinese Han population. Mol Psychiatry. 2021;26;4475–85.
Daugherty M, Polanuyer B, Farrell M, Scholle M, Lykidis A, de Crécy-Lagard V, et al. Complete reconstitution of the human coenzyme A biosynthetic pathway via comparative genomics. J Biol Chem. 2002;277:21431–9.
Cuenod M, Steullet P, Cabungcal J-H, Dwir D, Khadimallah I, Klauser P, et al. Caught in vicious circles: a perspective on dynamic feed-forward loops driving oxidative stress in schizophrenia. Mol Psychiatry. 2022;27:1886–97.
Zhang L, Silva TC, Young JI, Gomez L, Schmidt MA, Hamilton-Nelson KL, et al. Epigenome-wide meta-analysis of DNA methylation differences in prefrontal cortex implicates the immune processes in Alzheimer’s disease. Nat Commun. 2020;11:1–13.
Zhu L, Li Y, Xie X, Zhou X, Gu M, Jie Z, et al. TBKBP1 and TBK1 form a growth factor signalling axis mediating immunosuppression and tumourigenesis. Nat Cell Biol. 2019;21:1604–14.
Broce I, Karch CM, Wen N, Fan CC, Wang Y, Hong Tan C, et al. Immune-related genetic enrichment in frontotemporal dementia: an analysis of genome-wide association studies. PLoS Med. 2018;15:e1002487.
Mazza MG, Lucchi S, Rossetti A, Clerici M. Neutrophil-lymphocyte ratio, monocyte-lymphocyte ratio and platelet-lymphocyte ratio in non-affective psychosis: A meta-analysis and systematic review. World J Biol Psychiatry. 2020;21:326–38.
Birnbaum R, Weinberger DR. A genetics perspective on the role of the (neuro) immune system in schizophrenia. Schizophrenia Res. 2020;217:105–13.
Montano C, Taub MA, Jaffe A, Briem E, Feinberg JI, Trygvadottir R, et al. Association of DNA Methylation Differences With Schizophrenia in an Epigenome-Wide Association Study. JAMA Psychiatry. 2016;73:506–14.
Aberg KA, McClay JL, Nerella S, Clark S, Kumar G, Chen W, et al. Methylome-wide association study of schizophrenia: identifying blood biomarker signatures of environmental insults. JAMA Psychiatry. 2014;71:255–64.
Hannon E, Dempster E, Viana J, Burrage J, Smith AR, Macdonald R, et al. An integrated genetic-epigenetic analysis of schizophrenia: evidence for co-localization of genetic associations and differential DNA methylation. Genome Biol. 2016;17:176.
Liu J, Chen J, Ehrlich S, Walton E, White T, Perrone-Bizzozero N, et al. Methylation patterns in whole blood correlate with symptoms in Schizophrenia patients. Schizophrenia Bull. 2014;40:769–76.
Prados J, Stenz L, Courtet P, Prada P, Nicastro R, Adouan W. Borderline personality disorder and childhood maltreatment: a genome-wide methylation analysis. Genes Brain Behav. 2015;14:177–88.
Uddin M, Aiello AE, Wildman DE, Koenen KC, Pawelec G, de Los Santos R, et al. Epigenetic and immune function profiles associated with posttraumatic stress disorder. Proc Natl Acad Sci. 2010;107:9470–5.
Arranz MJ, Gallego-Fabrega C, Martín-Blanco A, Soler J, Elices M, Dominguez-Clavé E, et al. A genome-wide methylation study reveals X chromosome and childhood trauma methylation alterations associated with borderline personality disorder. Transl Psychiatry 2021;11:5.
Yang BZ, Zhang H, Ge W, Weder N, Douglas-Palumberi H, Perepletchikova F, et al. Child abuse and epigenetic mechanisms of disease risk. Am J Prev Med. 2013;44:101–7.
Sleiman P, Wang D, Glessner J, Hadley D, Gur RE, Cohen N, et al. GWAS meta analysis identifies TSNARE1 as a novel Schizophrenia / Bipolar susceptibility locus. Sci Rep. 2013;3:3075.
Li M, Shen L, Chen L, Huai C, Huang H, Wu X, et al. Novel genetic susceptibility loci identified by family based whole exome sequencing in Han Chinese schizophrenia patients. Transl Psychiatry. 2020;10:5.
Schrode N, Ho SM, Yamamuro K, Dobbyn A, Huckins L, Matos MR, et al. Synergistic effects of common schizophrenia risk variants. Nat Genet. 2019;51:1475–85.
Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19:1442–53.
Plooster M, Rossi G, Farrell MS, McAfee JC, Bell JL, Ye M, et al. Schizophrenia-Linked Protein tSNARE1 Regulates Endosomal Trafficking in Cortical Neurons. J Neurosci. 2021;41:9466–81.
Lopez-Lengowski K, Kathuria A, Gerlovin K, Karmacharya R. Co-Culturing Microglia and Cortical Neurons Differentiated from Human Induced Pluripotent Stem Cells. J Vis Exp. 2021. https://doi.org/10.3791/62480.
Ackerman SD, Luo R, Poitelon Y, Mogha A, Harty BL, D’Rozario M, et al. GPR56/ADGRG1 regulates development and maintenance of peripheral myelin. J Exp Med. 2018;215:941–61.
Chiou B, Gao C, Giera S, Folts CJ, Kishore P, Yu D, et al. Cell type-specific evaluation of ADGRG1/GPR56 function in developmental central nervous system myelination. Glia. 2021;69:413–23.
Millar MW, Corson N, Xu L. The Adhesion G-Protein-Coupled Receptor, GPR56/ADGRG1, Inhibits Cell-Extracellular Matrix Signaling to Prevent Metastatic Melanoma Growth. Front Oncol. 2018;8:8.
Olaniru OE, Pingitore A, Giera S, Piao X, Castañera González R, Jones PM, et al. The adhesion receptor GPR56 is activated by extracellular matrix collagen III to improve β-cell function. Cell Mol Life Sci. 2018;75:4007–19.
Tavares R, Wajnberg G, Scherer NM, Pauletti BA, Cassoli JS, Ferreira CG, et al. Unveiling alterative splice diversity from human oligodendrocyte proteome data. J Proteom. 2017;151:293–301.
Monin A, Baumann P, Griffa A, Xin L, Mekle R, Fournier M, et al. Glutathione deficit impairs myelin maturation: relevance for white matter integrity in schizophrenia patients. Mol Psychiatry. 2015;20:827.
Cabungcal JH, Steullet P, Morishita H, Kraftsik R, Cuenod M, Hensch TK, et al. Perineuronal nets protect fast-spiking interneurons against oxidative stress. Proc Natl Acad Sci USA. 2013;110:9130–5.
Kolomeets NS, Uranova NA Reduced number of satellite oligodendrocytes of pyramidal neurons in layer 5 of the prefrontal cortex in schizophrenia. Eur Arch Psychiatry Clin Neurosci. 2022;272:947–55.
Kebir O, Chaumette B, Rivollier F, Miozzo F, Lemieux Perreault LP, Barhdadi A, et al. Methylomic changes during conversion to psychosis. Mol Psychiatry. 2017;22:512–8.
Grinchii D, Dremencov E. Mechanism of action of atypical antipsychotic drugs in mood disorders. Int J Mol Sci. 2020;21:9532.
Hu W, Chen Z. The roles of histamine and its receptor ligands in central nervous system disorders: An update. Pharmacol Therap. 2017;175:116–32.
Karlstedt K, Senkas A, Åhman M, Panula P. Regional expression of the histamine H2 receptor in adult and developing rat brain. Neuroscience. 2001;102:201–8.
Haas HL, Sergeeva OA, Selbach O. Histamine in the nervous system. Physiol Rev. 2008;88:1183–1241.
Dai H, Kaneko K, Kato H, Fujii S, Jing Y, Xu A, et al. Selective cognitive dysfunction in mice lacking histamine H1 and H2 receptors. Neurosci Res. 2007;57:306–13.
Ogawa S, Yanai K, Watanabe T, Wang Z-M, Akaike H, Ito Y, et al. Histamine responses of large neostriatal interneurons in histamine H1 and H2 receptor knock-out mice. Brain Res Bull. 2009;78:189–94.
Iwabuchi K, Kubota Y, Ito C, Watanabe T, Watanabe T, Yanai K. Methamphetamine and brain histamine: A study using histamine‐related gene knockout mice. Ann N. Y Acad Sci. 2004;1025:129–34.
Mobarakeh JI, Takahashi K, Sakurada S, Kuramasu A, Yanai K. Enhanced antinociceptive effects of morphine in histamine H2 receptor gene knockout mice. Neuropharmacology. 2006;51:612–22.
Monette J, Mogun H, Bohn RL, Avorn J. Concurrent use of antiulcerative agents. J Clin Gastroenterol. 1997;24:207–13.
Kaminsky R, Moriarty T, Bodine J, Wolf D, Davidson M. Effect of famotidine on deficit symptoms of schizophrenia. Lancet. 1990;335:1351–2.
Assunção SSM, Ruschel SI, Rosa LDCR, Campos JAO, Alves MJO, Bracco OL, et al. Weight gain management in patients with schizophrenia during treatment with olanzapine in association with nizatidine. Braz J Psychiatry. 2006;28:270–6.
Farzin D, Hosseini SH, Shafaat A. A randomized double blind clinical trial in famotidine adjuvant therapy in schizophrenia. Iranian J Med Sci. 2005;30.
Meskanen K, Ekelund H, Laitinen J, Neuvonen PJ, Haukka J, Panula P, et al. A randomized clinical trial of histamine 2 receptor antagonism in treatment-resistant schizophrenia. J Clin Psychopharmacol. 2013;33:472–8.
Poyurovsky M, Tal V, Maayan R, Gil-Ad I, Fuchs C, Weizman A. The effect of famotidine addition on olanzapine-induced weight gain in first-episode schizophrenia patients: a double-blind placebo-controlled pilot study. Eur Neuropsychopharmacol. 2004;14:332–6.
Atmaca M, Kuloglu M, Tezcan E, Ustundag B. Nizatidine treatment and its relationship with leptin levels in patients with olanzapine‐induced weight gain. Hum Psychopharmacol: Clin Exp. 2003;18:457–61.
Atmaca M, Kuloglu M, Tezcan E, Ustundag B, Kilic N. Nizatidine for the treatment of patients with quetiapine‐induced weight gain. Hum Psychopharmacol: Clin Exp. 2004;19:37–40.
Orange P, Heath P, Wright S, Ramchand C, Kolkeiwicz L, Pearson R. Individuals with schizophrenia have an increased incidence of the H2R649G allele for the histamine H2 receptor gene. Mol Psychiatry. 1996;1:466–9.
Orange PR, Heath PR, Wright SR, Pearson R. Allelic variations of the human histamine H2 receptor gene. Neuroreport. 1996;7:1293–6.
Unnikrishnan A, Freeman WM, Jackson J, Wren JD, Porter H, Richardson A. The role of DNA methylation in epigenetics of aging. Pharmacol therapeutics. 2019;195:172–85.
Elliott HR, Tillin T, McArdle WL, Ho K, Duggirala A, Frayling TM, et al. Differences in smoking associated DNA methylation patterns in South Asians and Europeans. Clin Epigenetics. 2014;6:4.
Ji H, Ehrlich LI, Seita J, Murakami P, Doi A, Lindau P, et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467:338–42.
Razin A, Cedar H. DNA methylation and gene expression. Microbiol Rev. 1991;55:451–8.
Hannon E, Lunnon K, Schalkwyk L, Mill J. Interindividual methylomic variation across blood, cortex, and cerebellum: implications for epigenetic studies of neurological and neuropsychiatric phenotypes. Epigenetics. 2015;10:1024–32.
Acknowledgements
We thank all the contributors to the EU-GEI (WP2 group) study for their hard work: Kathryn Hubbard, Stephanie Beards, Simona A. Stilo, Mara Parellada, Pedro Cuadrado, José Juan Rodríguez Solano, Angel Carracedo, David Fraguas, Álvaro Andreu-Bernabeu, Gonzalo López, Bibiana Cabrera, Esther Lorente-Rovira, Paz Garcia-Portilla, Javier Costas, Estela Jiménez-López, Mario Matteis, Marta Rapado-Castro, Emiliano González, Covadonga M. Díaz-Caneja, Emilio Sánchez, Manuel Durán-Cutilla, Nathalie Franke, Fabian Termorshuizen, Daniella van Dam, Elsje van der Ven, Elles Messchaart, Marion Leboyer, Franck Schürhoff, Stéphane Jamain, Grégoire Baudin, Aziz Ferchiou, Baptiste Pignon, Jean-Romain Richard, Thomas Charpeaud, Anne-Marie Tronche, Flora Frijda, Giovanna Marrazzo, Crocettarachele Sartorio, Fabio Seminerio, Camila Marcelino Loureiro, Rosana Shuhama, Mirella Ruggeri, Chiara Bonetto, Doriana Cristofalo, Domnico Berardi, Marco Seri, Elena Bonora, Giuseppe D’Andrea, Laura Ferraro, Giada Tripoli, Silvia Amoretti, Gisela Mezquida. We thank strongly thank Romayne Gadelrab for her help with Fig. 3. We thank our funding bodies; The EU-GEI Project is funded by the European Community’s Seventh Framework Programme under grant agreement No. HEALTH-F2-2010-241909 (Project EU-GEI); Craig Morgan is part funded by the ESRC (ESRC Centre for Society and Mental Health at King’s College London: ESRC Reference: ES/S012567/1), and the NIHR Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London.
Author information
Authors and Affiliations
Contributions
LA conceptualised and designed the study, conducted the statistical analyses and wrote the initial draft; CW and RM conceptualised and designed the study; reviewed the initial draft and supervised the project; LZ created the DACT and conducted statistical analyses and reviewed the initial draft; PCS, MDF contributed to the design and reviewed the initial draft; MA, GT, VR, SAS, RK, CA, MA, MB, JB, LDH, CMD, CGA, LS, PBJ, HEJ, JBK, CLC, AL, ST, PML, PRM, JVO, BPR, JLS, JS, JPS, AS, IT, AT, EV, CM, ED reviewed the first draft and contributed to data collection and EUGEI planning, DD, AA reviewed the first draft, JM, EH, JB, ED reviewed the first draft and contributed to data collection and preparation; DQ data management.
Corresponding author
Ethics declarations
Competing interests
MB has been a consultant for, received grant/research support and honoraria from, and been on the speakers/advisory board of AB-Biotics, Adamed, Angelini, Casen Recordati, Janssen-Cilag, Menarini, Rovi and Takeda. CA has been a consultant to or has received honoraria or grants from Acadia, Angelini, Gedeon Richter, Janssen Cilag, Lundbeck, Minerva, Otsuka, Roche, Sage, Servier, Shire, Schering Plough, Sumitomo Dainippon Pharma, Sunovion and Takeda. PBJ declare to have consulted for Ricordati and Janssen. RM has received payments for non-promotional seminars from JANSSEN, SUNOVIAN, LUNDBECK AND OTSUKA.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Alameda, L., Liu, Z., Sham, P.C. et al. Exploring the mediation of DNA methylation across the epigenome between childhood adversity and First Episode of Psychosis—findings from the EU-GEI study. Mol Psychiatry 28, 2095–2106 (2023). https://doi.org/10.1038/s41380-023-02044-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-023-02044-9