Abstract
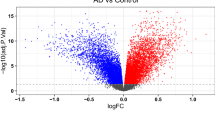
Alzheimer’s disease (AD) results in progressive cognitive decline owing to the accumulation of amyloid plaques and hyperphosphorylated tau. MicroRNAs (miRNAs) have attracted attention as a putative diagnostic and therapeutic target for neurodegenerative diseases. However, existing meta-analyses on AD and its association with miRNAs have produced inconsistent results. The primary objective of this study is to evaluate the magnitude and consistency of differences in miRNA levels between AD patients, mild cognitive impairment (MCI) patients and healthy controls (HC). Articles investigating miRNA levels in blood, brain tissue, or cerebrospinal fluid (CSF) of AD and MCI patients versus HC were systematically searched in PubMed/Medline from inception to February 16th, 2021. Fixed- and random-effects meta-analyses were complemented with the I2 statistic to measure the heterogeneity, assessment of publication bias, sensitivity subgroup analyses (AD severity, brain region, post-mortem versus ante-mortem specimen for CSF and type of analysis used to quantify miRNA) and functional enrichment pathway analysis. Of the 1512 miRNAs included in 61 articles, 425 meta-analyses were performed on 334 miRNAs. Fifty-six miRNAs were significantly upregulated (n = 40) or downregulated (n = 16) in AD versus HC and all five miRNAs were significantly upregulated in MCI versus HC. Functional enrichment analysis confirmed that pathways related to apoptosis, immune response and inflammation were statistically enriched with upregulated pathways in participants with AD relative to HC. This study confirms that miRNAs’ expression is altered in AD and MCI compared to HC. These findings open new diagnostic and therapeutic perspectives for this disorder.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
08 April 2022
A Correction to this paper has been published: https://doi.org/10.1038/s41380-022-01534-6
References
Prince M, Ali GC, Guerchet M, Prina AM, Albanese E, Wu YT. Recent global trends in the prevalence and incidence of dementia, and survival with dementia. Alzheimers Res Ther. 2016;8:23.
Checkoway H, Lundin JI, Kelada SN Neurodegenerative diseases. IARC Sci Publ. 2011:407-19.
Alzheimer’s A. 2016 Alzheimer’s disease facts and figures. Alzheimers Dement. 2016;12:459–509.
Kumar A, Nisha CM, Silakari C, Sharma I, Anusha K, Gupta N, et al. Current and novel therapeutic molecules and targets in Alzheimer’s disease. J Formos Med Assoc. 2016;115:3–10.
Salloway S, Sperling R, Fox NC, Blennow K, Klunk W, Raskind M, et al. Two phase 3 trials of bapineuzumab in mild-to-moderate Alzheimer’s disease. N Engl J Med. 2014;370:322–33.
Huang LK, Chao SP, Hu CJ. Clinical trials of new drugs for Alzheimer disease. J Biomed Sci. 2020;27:18.
Brosius J. Waste not, want not-transcript excess in multicellular eukaryotes. Trends Genet. 2005;21:287–8.
Junn E, Mouradian MM. MicroRNAs in neurodegenerative diseases and their therapeutic potential. Pharm Ther. 2012;133:142–50.
Qureshi IA, Mehler MF. Non-coding RNA networks underlying cognitive disorders across the lifespan. Trends Mol Med. 2011;17:337–46.
Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, et al. A uniform system for microRNA annotation. RNA. 2003;9:277–9.
Jiang W, Zhang Y, Meng F, Lian B, Chen X, Yu X, et al. Identification of active transcription factor and miRNA regulatory pathways in Alzheimer’s disease. Bioinformatics. 2013;29:2596–602.
Wang J, Chen C, Zhang Y. An investigation of microRNA-103 and microRNA-107 as potential blood-based biomarkers for disease risk and progression of Alzheimer’s disease. J Clin Lab Anal. 2020;34:e23006.
Hu YB, Li CB, Song N, Zou Y, Chen SD, Ren RJ, et al. Diagnostic value of microRNA for Alzheimer’s disease: a systematic review and meta-analysis. Front Aging Neurosci. 2016;8:13.
Boissonneault V, Plante I, Rivest S, Provost P. MicroRNA-298 and MicroRNA-328 regulate expression of mouse beta-amyloid precursor protein-converting enzyme 1. J Biol Chem. 2009;284:1971–81.
Wang X, Liu D, Huang H-Z, Wang Z-H, Hou T-Y, Yang X, et al. A novel MicroRNA-124/PTPN1 signal pathway mediates synaptic and memory deficits in Alzheimer’s disease. Biol Psychiatry. 2018;83:395–405.
McKeever PM, Schneider R, Taghdiri F, Weichert A, Multani N, Brown RA, et al. MicroRNA expression levels are altered in the cerebrospinal fluid of patients with young-onset Alzheimer’s disease. Mol Neurobiol. 2018;55:8826–41.
Kiko T, Nakagawa K, Tsuduki T, Furukawa K, Arai H, Miyazawa T. MicroRNAs in plasma and cerebrospinal fluid as potential markers for Alzheimer’s disease. J Alzheimers Dis. 2014;39:253–9.
Takousis P, Sadlon A, Schulz J, Wohlers I, Dobricic V, Middleton L, et al. Differential expression of microRNAs in Alzheimer’s disease brain, blood, and cerebrospinal fluid. Alzheimers Dement. 2019;15:1468–77.
Brain, blood, and cerebrospinal fluid microRNAs and noncoding RNAs in Alzheimer’s disease: a systematic review and meta-analysis. https://www.crd.york.ac.uk/prospero/display_record.php?ID=CRD42020150993, 2020, Accessed Date Accessed 2020 Accessed.
Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6:e1000097.
Cochrane Handbook for Systematic Reviews of Interventions version 6.0. http://www.training.cochrane.org/handbook, 2019, Accessed Date Accessed 2019 Accessed.
The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp, 2012, Accessed Date Accessed 2012 Accessed.
Li J, Han X, Wan Y, Zhang S, Zhao Y, Fan R, et al. TAM 2.0: tool for MicroRNA set analysis. Nucleic Acids Res. 2018;46:W180–85.
Idda ML, Munk R, Abdelmohsen K, Gorospe M. Noncoding RNAs in Alzheimer’s disease. Wiley Interdiscip Rev RNA. 2018;9:e1463.
Millan MJ. Linking deregulation of non-coding RNA to the core pathophysiology of Alzheimer’s disease: An integrative review. Prog Neurobiol. 2017;156:1–68.
Kim C, Kang D, Lee EK, Lee JS. Long noncoding RNAs and RNA-binding proteins in oxidative stress, cellular senescence, and age-related diseases. Oxid Med Cell Longev. 2017;2017:2062384.
Zhu Y, Wang L, Yin Y, Yang E. Systematic analysis of gene expression patterns associated with postmortem interval in human tissues. Sci Rep. 2017;7:5435.
Vella LJ, Greenwood DL, Cappai R, Scheerlinck JP, Hill AF. Enrichment of prion protein in exosomes derived from ovine cerebral spinal fluid. Vet Immunol Immunopathol. 2008;124:385–93.
Cuk K, Zucknick M, Heil J, Madhavan D, Schott S, Turchinovich A, et al. Circulating microRNAs in plasma as early detection markers for breast cancer. Int J Cancer. 2013;132:1602–12.
Mizuno H, Nakamura A, Aoki Y, Ito N, Kishi S, Yamamoto K, et al. Identification of muscle-specific microRNAs in serum of muscular dystrophy animal models: promising novel blood-based markers for muscular dystrophy. PLoS One. 2011;6:e18388.
Pigati L, Yaddanapudi SC, Iyengar R, Kim DJ, Hearn SA, Danforth D, et al. Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS One. 2010;5:e13515.
Jiang MC, Ni JJ, Cui WY, Wang BY, Zhuo W. Emerging roles of lncRNA in cancer and therapeutic opportunities. Am J Cancer Res. 2019;9:1354–66.
Knebl J, DeFazio P, Clearfield MB, Little L, McConathy WJ, McPherson R, et al. Plasma lipids and cholesterol esterification in Alzheimer’s disease. Mech Ageing Dev. 1994;73:69–77.
Ginsberg L, Atack JR, Rapoport SI, Gershfeld NL. Regional specificity of membrane instability in Alzheimer’s disease brain. Brain Res. 1993;615:355–7.
Chang TY, Chang C. ApoE and lipid homeostasis in Alzheimer’s disease: introduction to the thematic review series. J Lipid Res. 2017;58:823.
Kitagishi Y, Nakanishi A, Ogura Y, Matsuda S. Dietary regulation of PI3K/AKT/GSK-3beta pathway in Alzheimer’s disease. Alzheimers Res Ther. 2014;6:35.
Roth KA. Caspases, apoptosis, and Alzheimer disease: causation, correlation, and confusion. J Neuropathol Exp Neurol. 2001;60:829–38.
Kumar A, Singh A. Ekavali. A review on Alzheimer’s disease pathophysiology and its management: an update. Pharmacol Rep. 2015;67:195–203.
Hardy J. The amyloid hypothesis for Alzheimer’s disease: a critical reappraisal. J Neurochem. 2009;110:1129–34.
Rosenmann H. Immunotherapy for targeting tau pathology in Alzheimer’s disease and tauopathies. Curr Alzheimer Res. 2013;10:217–28.
dos Santos P, Leide C, Ozela PF, de Fatima de Brito Brito M, Pinheiro AA, Padilha EC, et al. Alzheimer’s disease: a review from the pathophysiology to diagnosis, new perspectives for pharmacological treatment. Curr medicinal Chem. 2018;25:3141–59.
Choi S-H, Aid S, Caracciolo L, Minami SS, Niikura T, Matsuoka Y, et al. Cyclooxygenase-1 inhibition reduces amyloid pathology and improves memory deficits in a mouse model of Alzheimer’s disease. J neurochemistry. 2013;124:59–68.
Rice DB, Kloda LA, Levis B, Qi B, Kingsland E, Thombs BD. Are MEDLINE searches sufficient for systematic reviews and meta-analyses of the diagnostic accuracy of depression screening tools? A review of meta-analyses. J Psychosom Res. 2016;87:7–13.
Marshall IJ, Marshall R, Wallace BC, Brassey J, Thomas J. Rapid reviews may produce different results to systematic reviews: a meta-epidemiological study. J Clin Epidemiol. 2019;109:30–41.
Acknowledgements
This paper demonstrates independent research and all authors are acknowledged for their contributions to this study.
Author information
Authors and Affiliations
Contributions
SY and JIS designed the study. SY, SUK and JIS searched the literature, and extracted data. Any discrepancies were resolved via discussion between SY, SUK and JIS. SY, SUK, YK, GHJ, KHL and JIS undertook the statistical analyses and interpreted the data. SY and SUK made the figures and tables. All authors drafted and critically revised the manuscript. All authors approved the final version of the manuscript for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: In this article the affiliation details for Author JAE IL SHIN were incorrectly given as ‘Department of Psychiatry, Yonsei University Wonju College of Medicine, Wonju, Republic of Korea’ but should have been ‘Department of Pediatrics, Yonsei University College of Medicine, Seoul, Republic of Korea’.
Supplementary information
Rights and permissions
About this article
Cite this article
Yoon, S., Kim, S.E., Ko, Y. et al. Differential expression of MicroRNAs in Alzheimer’s disease: a systematic review and meta-analysis. Mol Psychiatry 27, 2405–2413 (2022). https://doi.org/10.1038/s41380-022-01476-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-022-01476-z
This article is cited by
-
The role of microRNAs in understanding sex-based differences in Alzheimer’s disease
Biology of Sex Differences (2024)
-
The GABAergic system in Alzheimer’s disease: a systematic review with meta-analysis
Molecular Psychiatry (2023)
-
Dissecting early life stress-induced adolescent depression through epigenomic approach
Molecular Psychiatry (2023)