Abstract
An anxious or inhibited temperament (IT) early in life is a major risk factor for the later development of stress-related psychopathology. Starting in infancy, nonhuman primates, like humans, begin to reveal their temperament when exposed to novel situations. Here, in Study 1 we demonstrate this infant IT predicts adult behavior. Specifically, in over 600 monkeys, we found that individuals scored as inhibited during infancy were more likely to refuse treats offered by potentially-threatening human experimenters as adults. In Study 2, using a sample of over 4000 monkeys from a large multi-generational family pedigree, we demonstrate that infant IT is partially heritable. The data revealed infant IT to reflect a co-inherited substrate that manifests across multiple latent variables. Finally, in Study 3 we performed whole-genome sequencing in 106 monkeys to identify IT-associated single-nucleotide variations (SNVs). Results demonstrated a genome-wide significant SNV near CTNNA2, suggesting a molecular target worthy of additional investigation. Moreover, we observed lower p values in genes implicated in human association studies of neuroticism and depression. Together, these data demonstrate the utility of our model of infant inhibited temperament in the rhesus monkey to facilitate discovery of genes that are relevant to the long-term inherited risk to develop anxiety and depressive disorders.
Similar content being viewed by others
Introduction
An anxious temperament (AT) or inhibited temperament (IT) early in life is a major risk factor for the later development of anxiety and depressive disorders [1,2,3,4]. Children with an extreme IT early in life are characterized by increased behavioral inhibition and anxiety in novel and potentially threatening contexts. In humans, IT reflects an inhibited nature or disposition that can define how an individual approaches novel situations throughout their life [5,6,7]. Although IT often emerges during the second year of life around the time that a child develops the ability to behaviorally cope with threat [8, 9] it can be preceded by increased reactivity in infancy [5]. Temperament is partially inherited, and in human samples the heritability estimates range from 20 to 50% [10,11,12,13,14,15,16]. Because IT is heritable, and a risk-factor for the later development of stress-related psychopathology, IT likely reflects an underlying genetic disposition and/or mediates the relationship between genetic variation and psychopathology.
To understand which molecular components contribute to anxiety and depressive disorders, researchers have begun to perform genome-wide association studies (GWAS) with dispositional anxiety and psychopathology in humans. Large-scale efforts for anxiety disorders [17, 18], depressive disorders [19, 20], and neuroticism [21] are all ongoing. Polygenic risk-scores and genetic correlation analyses, suggest each of these disorders partially overlap in their genetic substrates [17, 21]. Although some studies have identified genes with significant single-nucleotide variants (SNVs; e.g., in CRHR1, ESR1, NTRK2, etc.) [17, 21, 22], the specific SNVs influencing psychopathology may not be the same in individuals with different genetic backgrounds and/or who were exposed to different environmental stressors.
Rhesus macaques provide a powerful and well-validated animal model to study the genetic and molecular bases of early-life dispositional anxiety because of their similarity to humans. Next to humans, rhesus macaques are the most widely geographically distributed primates in the world, thereby exhibiting outstanding ecological flexibility and adaptability [23, 24]. As Old World monkeys, rhesus macaques are phylogenetically close to humans (with a common ancestor ~10–12 million years closer to humans than marmosets and ~50 million years closer than mice). This recent evolutionary divergence between rhesus macaques and humans is reflected in similarities in their genomes, transcript expression, brain circuits, and resulting socio-emotional behavior [25,26,27,28]. Thus, the rhesus monkey model provides a unique opportunity for translational research that supports and extends the insights gained from human genetic studies.
Capitanio et al. have developed and implemented a rhesus monkey model of infant IT, as part of their standardized BioBehavioral Assessment (BBA) that has been applied to over 4000 animals. Infant IT is characterized by below-average expression of latent variables which emerge from behavioral observations spread over 2 days, termed “Activity” and “Emotionality”, both of which are indicative of behavioral inhibition [29](see methods for details). Notably, molecular studies of dispositional anxiety in macaques are concordant with human GWAS studies, hinting at evolutionarily-conserved mechanisms (e.g., CRH [30, 31], and neurotrophic pathways [4, 32, 33]. Infant IT among macaques provides an excellent model for the study of the human risk for anxiety and depressive disorders [4, 34].
The rhesus monkey model of IT provides numerous advantages that facilitate the identification of genes and molecules that contribute to the risk for psychopathology. Rhesus populations have higher levels of genetic variation and lower linkage disequilibrium than do equivalent sample sizes from human populations, likely due to the historic population bottleneck experienced by ancient human populations that gave rise to modern humans [26, 35,36,37,38]. Importantly, this genetic variation is demonstrated in the 853 rhesus macaque whole-genome sequences that Dr. Rogers et al. have collected from US primate centers, which has identified >85 million SNVs, including 408,496 missense variants, 9921 stop codons gained and 7918 splice acceptor or donor variants [39]. Moreover, the breeding strategy at primate centers results in large multi-generational pedigrees in which each animal has many close and more distant relatives, and this will enrich the population for putatively “rare” variation, thus increasing statistical power in heritability and genetic associations analyses [40].
Here, we demonstrate that infant IT in macaques can be used as an endophenotype to complement human studies of psychopathology, and gain insight into the nature of inherited anxiety. We demonstrate that infant IT is associated with lasting behavioral changes (study 1); show that infant IT is heritable (study 2); and begin to study the genetic variation that underlie this early-life risk-factor (study 3).
Method
Methods overview
All studies were performed in rhesus macaques (Macaca mulatta) in accordance with the federal guidelines of animal use and care and with the approval of the University of California, Davis Institutional Animal Care and Use Committee. Primary analyses across all studies included animals that underwent BBA-testing during infancy (3–4 months of age). Subsets of animals were selected for Food Retrieval Task testing in Study 1 (n = 679; 59M/620F), heritability analyses in Study 2 (n = 4433; 2019M/2414F), and whole-genome sequencing in Study 3 (n = 106; 49M/57F). The only animals that did not undergo BBA-testing were a subset of female animals that underwent test-retest analyses in Study 1 (>2 tests: n = 649; >3 tests: n = 288; 4 tests: n = 88). Additional detailed methods can be found in the Supplementary Methods as well as previous publications [29].
Assessment of infant IT (Studies 1–3)
The methods for scoring IT in the BBA program have been previously described [29, 34]. Animals (90–120 days old) were relocated for a 25 h testing period (see supplementary methods). IT is defined based on four factors that emerged from factor analyses in a subset of several hundred animals [29]. Animals were considered to be “inhibited” if their scores were below the mean for both “Activity” and “Emotionality” across 2 days of testing, otherwise they were classified as “not inhibited.” The “Activity” factor includes time locomoting; time NOT hanging from the top or side of the cage; rate of environmental exploration; and whether or not the animals ate food*, drank water*, or were seen crouching in the cage* (* = dichotomized due to rarity). The “Emotionality” factor includes rate of cooing; rate of barking; and whether the animals scratched*, displayed threats*, or lipsmacked* (* = dichotomized due to rarity).
Food retrieval task
The Food Retrieval task was administered at ~6:00 am on the day after relocation, prior to morning health and husbandry, by a technician that was blind to infant IT scores. To ensure there was no familiarity with the testing context, the Food Retrieval Task was performed in a different location than BBA-testing. The technician stood in front of the cage, then approached the animal and hand presented a food treat for 5 s, taking care to avert her eyes from the monkey (a stopwatch with an audible beep was used for timing). Treat retrieval was recorded. If the animal did not accept the treat, the technician placed the treat on the forage board and stepped back from the cage, averted her eyes, waited 5 s, and again recorded whether or not the treat was retrieved. Three trials were run consecutively for each animal. Because humans in such close proximity can be perceived as threatening, the Food Retrieval task sets up a potential conflict for the animals between a fear of the human versus attraction to a favored treat.
To estimate the relationship between infant IT and refusal to take food in the food retrieval task, we used logistic regressions. To estimate test-retest stability of the dichotomous reach variable, we used chi-squre tests [41, 42]. All statistical analyses were implemented in Python (version 3.7.3), statsmodels (version 0.10.0; https://www.statsmodels.org/stable/index.html[43]; was used for regression analyses, and (Pingouin; https://pingouin-stats.org/[44]; was used to perform chi-squared tests.
Heritability of IT
For this study, we analyzed variation among 4433 infants assessed through the BBA program between May of 2001 and January of 2017. The total sample consisted of 407 inhibited females, 403 inhibited males, 2007 non-inhibited females, and 1616 non-inhibited males. As in our previous work [45,46,47,48], all heritability and co-heritability estimates were performed using SOLAR-Eclipse (http://solar-eclipse-genetics.org/). Prior to heritability estimation phenotype variables were normalized using an inverse normal transformation. All heritability analyses controlled for sex. Because all animals were assessed between 3 and 4 months of age, we did not include Age or Age-squared as covariates in heritability analyses.
Methods for whole-genome sequencing and mapping
Blood samples were collected from 36 inhibited animals and 70 non-inhibited animals. DNA was extracted and sequenced at the Human Genome Sequencing Center, Baylor College of Medicine using either the Illumina HiSeq 2000 or Illumina HiSeq X Ten system. WGS sequence data for the 106 animals are publicly available through the NCBI SRA (https://www.ncbi.nlm.nih.gov/biosample/?term=Bio+Behavior+Assessment). Paired end reads were aligned to the rhesus Mmul_10 reference using BWA mem with an average mapped sequence depth of 33.66X across the samples. The GATK v. 4.1.2.0 [49] pipeline was used to identify single-nucleotide variants (SNVs) and insertions/deletions (indels) smaller than 7bp. Variant Effect Predictor [50] was used to annotate variants based on merged Ensembl and RefSeq gene models.
IT-related variants were analyzed using FaST-LMM [51], which implements a linear mixed model that takes potential relatedness into account, controlling for sex. Sequence variants of interest were further examined by lifting the rhesus positions over to the orthologous human position and performing CADD analysis [52].
Permutation tests were used to compare IT-related associations to relevant gene lists extracted from published human genome-wide gene-association studies (GWGAS). First, the minimum p value for each gene was computed. Then the average minimum p value for IT-associations for each gene in the target-list was computed and stored. Finally, for each analysis we performed 10,000 permutations with a similarly sized set of randomly selected genes, and determined the average p value of those gene-sets. The p value was computed as the proportion of permutations that resulted in a lower p value than the target gene-set.
Results
Study 1: Infant IT predicts later-life behavior
To begin, we determined whether infant IT reflects an animal’s stable disposition throughout their lifespan. We assessed behavioral inhibition during a Food Retrieval Task, when adult animals (n = 679) were offered a treat by a human experimenter. We hypothesized that inhibited animals would forgo reward in the presence of this potentially threatening human. Logistic regressions demonstrated animals with high levels of IT at 3–4 months of age are less willing to take a treat years later (z = 3.248, p = 0.001; Fig. 1).
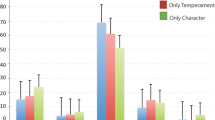
A Proportion of animals that refused treats in the Food Retrieval Task in relation to infant IT. B A histogram of ages for animals when tested in the Treat Refusal Task, on average >4 years after assessment of infant IT. C The factors that contribute to IT, i.e., Day 1 and 2 Activity and Emotionality, are each related to the propensity to refuse treats. Error-bars reflect bootstrapped 95% confidence interval.
IT is a composite measure of two latent factors, across 2 days, i.e., Day 1 and Day 2 Activity & Emotionality. To be sure each factor of IT was contributing to the underlying temperamental variable, we assessed each variable as a predictor of treat refusal. Analyses revealed each measure, Activity and Emotionality, on each day to be predictive of treat refusal (all p’s < 0.014). Supplementary analyses revealed no significant effects of sex (p = 0.209); significant effects of IT on treat refusal in both males (t = 2.710, p = 0.007) and females (t = 3.193, p = 0.001); an effect of age on treat refusal, such that older animals were less likely to refuse a treat (z = −5.419, p < 0.001); and that IT remained significantly associated with treat refusal while controlling for age (z = 3.785, p < 0.001) (see Supplementary Results for additional descriptions and consideration of IT as a continuous variable). Interestingly, although animals have multiple opportunities to retrieve treats, the Food Retrieval Task typically results in an all-or-nothing result, with only ~17% (49/292) of animals who did not retrieve a treat on the first trial going on to retrieve any treat. Unsurprisingly, we obtained similar results when examining first-treat refusal, for IT (z = 3.248 p = 0.001), age (z = −6.064, p < 0.001), and IT controlling for age (2.935, p = 0.003). Together these data suggest that IT as assessed in this protocol is a trait-like measure, which is susceptible to change with experience, but does remain detectably consistent within an animal across contexts as they mature.
We next evaluated treat-refusal as a stable assessment. In a separate group of 649 female animals, we performed the food retrieval task multiple times, longitudinally. Animals received 2 (N = 649) to 4 (N = 88) assessments, on average 1.8 years apart. Chi-squared tests revealed treat-inhibition to be stable between each assessment (p’s < 0.001; Fig. 2), such that 69% of animals responded consistently during the 4th assessment on average 5.24 years later (range = 1.9–9.2 years). Again, treat refusal tended to be all-or-nothing, with 82% of animals that refused the first treat refusing all treats (chi2 = 430.68, p < 0.000001), and first-treat retrieval showing similar stability (p’s < 0.001).
Together, these data demonstrate infant IT reflects life-long behavioral inhibition, that reflects varied behaviors across novel contexts.
Study 2: Infant IT is heritable
We next examined the heritability of IT, and its contributing factors, across 4433 animals that were part of a large multi-generational pedigree. Results showed that IT is significantly heritable with h2 = 0.19 (stdh2 = 0.027; p = 3.7e-27; Fig. 3A). Results also found IT’s latent factors, i.e., Day 1 and Day 2 Activity and Emotionality, to be significantly heritable (p’s < 1.0e-16) with h2 values ranging from 0.17 to 0.30. To test the extent to which the latent factors contributing to IT resulted from overlapping genetic variation, we performed genetic correlation analyses. Genetic correlation analyses test the extent to which covariation in phenotypes are associated with covariation in estimated genetic variation, as estimated by relatedness. Results demonstrated all latent factors that comprise IT were significantly genetically correlated with each other (rho-g estimates from 0.45 to 0.89; p’s < 0.0001) and with IT (rho-g estimates 0.65–0.90, p’s < 1 × 10−9; Fig. 3B). These data suggest a partially shared genetic substrate that contributes to these latent factors.
Inhibited Temperament (IT) is heritable, as are the contributing latent factors Activity and Emotionality across both days of testing (A) error-bars reflect standard deviation of the heritability estimate). In addition, IT shows a significant negative genetic correlation with each of latent factors, which in turn are positively correlated with each other (B).
Together, the data from Study 2 demonstrate infant IT is significantly heritable, making it an ideal starting point to identify genetic variation. Here, we focus genetic analyses on IT which is likely to identify genetic variation that contribute to multiple latent variables, and are less likely to be specifically related to an individual feature of IT (e.g., propensity toward locomotion).
Study 3: GWAS of IT
In this study, we surveyed BBA subjects for DNA sequence polymorphisms that might reasonably be hypothesized to influence IT, and determined whether the results were consistent with relevant human GWAS studies. Across the 106 individuals, we identified 53,030,128 SNVs and 6,435,882 indels using whole-genome sequencing, and used FaST-LMM to identify IT-related SNVs, taking relatedness into account. We identified a single SNV (13:27491805:C:T) that exceeded a significance threshold p < 5 × 10−8 which is a standard threshold for GWAS genome wide significance (Table S1, Fig. 4). No indels met this threshold. Two additional nearby SNVs had the third and fourth lowest p values in the dataset, but failed to reach the genome wide significance threshold (Table S1, Fig. 4B), including 13:27444729:G:C, 47,076bp upstream and 13:27493293:T:A, which was 1477bp downstream of the top SNV. All of these SNVs are intergenic, but 13:27491805:C:T is 473,868 bp downstream of the 3′ end of Catenin Alpha-2 (CTNNA2). However, although follow-up analyses revealed 4 indels and 43 SNVs in CTNNA2, we were unable to identify variation that was both related to IT (p < 0.05) and predicted to be functional (i.e., CADD > 10, see supplementary results, Tables S2–S3).
A Genome-wide Manhattan plot of BioBehavioral Assessment (BBA) associations with SNVs. The x-axis is the chromosomes and y-axis is the −log10 of the FaST-LMM p value. The genome-wide significance threshold p < 5 × 10−8 is represented by the red line. The significant SNV is highlighted in green. B A description of the significant SNV. C Chromosome 13 Manhattan plot of BioBehavioral Assessment (BBA) associations with SNVs. The x-axis is the chromosomes and y-axis is the −log10 of the FaST-LMM p value. The genome-wide significance threshold p < 5 × 10−8 is represented by the red line. The significant SNV is highlighted in green. D UCSC Genome Browser view of SNVs with FaST-LMM −log10 p values in the vicinity of CTNNA2. The genome-wide significance threshold p < 5 × 10−8 is represented by the red line.
In addition to our genome-wide significant hit in CTNNA2, we relaxed the formal genome-wide significance threshold and explored variation in other genes that were marginally significantly (p < 0.01) associated with IT (Table S4), but did not reach genome-wide significance. Interestingly, our results revealed uncorrected associations with genetic variation in genes we have previously implicated in inhibition, including an 3′ UTR variant in NTRK3 (p = 0.005) and an intronic variant in PRKCD (p = 0.007) [31, 33, 53]. Such associations must be considered less meaningful but can provide indications of genetic effects that deserve further study.
Using permutation analyses, we tested for IT-related enrichment in genes relevant to stress-related psychopathology to determine if further genetic studies of monkey IT are likely to identify evolutionarily-conserved genes and pathways that are relevant to human psychopathology. Because species differences prevent base-pair by base-pair comparisons, we examined gene-level enrichment based on GWGAS from studies of human Neuroticism (547 genes; Nagel) [21], Anxiety Disorders (31 genes; Levey) [17], and Depressive Disorders (251 genes; Coleman) [22]. We examined the minimum IT-related p value for target genes deemed significant in published GWGAS studies. Permutation analyses found the average minimum p value to be significantly lower in target gene lists as compared to randomly selected genes, for neuroticism (Nagel: p < 0.0001) and depressive disorders (Coleman: p = 0.0018), but not Anxiety Disorders (Levey: p = 0.21; though this list only contained 31 genes) (Fig. 5). Interestingly, genes that were significant in the human GWGASs and the current analyses (p < 0.005, uncorrected) include CTNNA2, PRKCD, NTRK3 and ESR1, highlighting the potential for identifying evolutionarily-conserved mechanisms. These data highlight the promise of our nonhuman primate approach, and suggest that additional data will identify additional IT-related genetic signals.
Permutation tests revealed the average minimum p value from genes found in human GWGAS studies of Neuroticism (p < 0.001) and Major Depressive Disorder (p = 0.011) was significantly lower than the average minimum p value in equally sized random subsets of genes. This same analysis did not reach significance for a GWGAS of Anxiety Disorders (p = 0.201).
Discussion
An early-life IT represents one of the strongest known risk-factors for the later development of psychopathology. Here, we demonstrate infant IT in rhesus macaques to be a heritable endophenotype that reflects stable context-independent behavior, and demonstrates promise for identifying molecules that contribute to stress-related psychopathology in humans. The data suggest infant IT is a potentially continuous measure that reflects underlying biological processes that manifest across different behaviors in different contexts (see Supplementary Results). These data mirror the work done in humans, in which children classified as behaviorally inhibited tend to remain inhibited as they mature, and are at a substantial risk to develop stress-related psychopathology [1,2,3, 54,55,56,57].
The extremely large number of phenotyped animals (N > 4000) that are part of a large multi-generational pedigree, provides compelling evidence for heritability, and a unique opportunity for further study of the specific SNVs that contribute to IT. Though generally consistent with our previous behavioral genetics studies of adolescent/adult rhesus macaques [30, 34, 36, 46, 58,59,60], previous heritability estimates tended to be slightly higher (e.g., ~25–40% heritable: [30, 45,46,47,48]. The analyses we describe here include over 4000 animals and found infant IT to be ~19% heritable (see Fig. 3). It is possible that these heritability estimates will increase as animals mature, as has been observed in twin studies of human anxiety [61, 62]. Like with human anxiety and depressive disorders, infant IT was associated with the family structure, such that inhibited animals are more likely to be related to inhibited animals.
Our preliminary genome-wide association analyses revealed a promising hit near the CTNNA2 gene in chromosome 13, which encodes Catenin alpha-2. This is of particular interest because CTNNA2 has been previously implicated in human association studies, including a GWGAS of anxiety disorders, and encodes a neuron-specific catenin that is important for cell-to-cell adhesion and synaptic plasticity, is expressed throughout cortical and subcortical structures, and is implicated in numerous studies of IT-related phenotypes [17, 63,64,65,66,67,68,69,70]. The association analysis presented here does not constitute evidence for a definitive association between CTNNA2 and infant IT in the rhesus monkey. That said, together with findings in humans, these data contribute to the rationale for further study of CTNNA2 in anxiety-related behavior in animal models, and highlight a potential molecular mechanism that may drive stable anxiety across the lifespan. More generally, our results are consistent with many molecules contributing to IT, reflecting disruption across many of the brain regions and cell-types that are thought to play a role in anxiety and threat-processing (See Supplementary discussion) [30, 32, 33, 46, 47, 53].
Moreover, these data suggest that further study of infant ITs genetics may identify molecules that are relevant to human psychopathology that can be targeted for mechanistic study. In this sample of 106 rhesus macaques, we identified IT-associations that were enriched for genes identified in GWGAS studies of ~449k subjects phenotype for neuroticism and ~624k subjects phenotyped for major depressive disorder in humans (including ~185k patients). We did not find significant enrichment for the genes implicated in a GWGAS of ~200k subjects phenotyped for anxiety disorders. This is likely because this study only identified 31 genes and will likely increase as the number of anxiety disorder genes goes up. Interestingly, although not significantly enriched for average p value, the CTNNA2 gene was identified in the million veterans analysis of anxiety disorders, suggesting CTNNA2 is a relevant gene, like CRHR1 [30], that is shared between human anxiety disorders and rhesus monkey infant-IT. Together, these data highlight the utility of using our rhesus monkey model of infant IT to identify molecules that may be relevant to human psychopathology, and suggest there is a shared molecular substrate that is conserved across primates.
Conclusion
Elucidating the genes and genetic mechanisms that predispose individuals to stress-related psychopathology can inform our understanding of psychopathology, and guide the development of biologically-informed treatments. Understanding of disease mechanisms and developing optimal treatments requires human epidemiological insights, as well as well-validated animal models. Future studies will be needed to overcome the limitations of the current preliminary GWAS study, and increase the number of subjects to obtain additional statistical power. Importantly, with the power afforded by a pedigree-based whole-genome sequencing approach in a genetically variable species, nonhuman primate studies may be able to identify genes and specific genetic variants that would otherwise take hundreds of thousands of subjects using a traditional GWAS approach [26, 36, 38, 40]. Here, we help provide support for a translational model that can support and extend the insights gained from human genetic studies to identify the mechanisms that underlie stress-related psychopathology.
References
Biederman J, Hirshfeld-Becker DR, Rosenbaum JF, Hérot C, Friedman D, Snidman N, et al. Further evidence of association between behavioral inhibition and social anxiety in children. Am J Psychiatry. 2001;158:1673–9.
Clauss JA, Blackford JU. Behavioral inhibition and risk for developing social anxiety disorder: a meta-analytic study. J Am Acad Child Adolesc Psychiatry. 2012;51:1066–75.e1.
Fox NA, Henderson HA, Marshall PJ, Nichols KE, Ghera MM. Behavioral inhibition: linking biology and behavior within a developmental framework. Annu Rev Psychol. 2005;56:235–62.
Fox AS, Kalin NH. A translational neuroscience approach to understanding the development of social anxiety disorder and its pathophysiology. Am J Psychiatry. 2014;171:1162–73.
Kagan J. Temperament and the reactions to unfamiliarity. Child Dev. 1997;68:139–43.
Kagan J, Reznick JS, Snidman N. Biological bases of childhood shyness. Science. 1988;240:167–71.
Zentner M, Shiner R. Fifty years of progress in temperament research: a synthesis of major themes, findings, challenges, and a look forward. In M. Zentner & R. L. Shiner editors, Handb Temperam. The Guilford Press (p. 673–700).
Scarr S, Salapatek P. Patterns of fear development during infancy. Merrill-Palmer Q Behav Dev. 1970;16:53–90.
Schaffer HR. The onset of fear of strangers and the incongruity hypothesis. J Child Psychol Psychiatry. 1966;7:95–106.
Dilalla LF, Kagan J, Reznick JS. Genetic etiology of behavioral inhibition among 2-year-old children. Infant Behav Dev. 1994;17:405–12.
Dunn J, Plomin R. Determinants of maternal behaviour towards 3-year-old siblings. Br J Dev Psychol. 1986;4:127–37.
Goldsmith HH, Gottesman II. Origins of variation in behavioral style: a longitudinal study of temperament in young twins. Child Dev. 1981;52:91–103.
Robinson JL, Kagan J, Reznick JS, Corley R. The heritability of inhibited and uninhibited behavior: a twin study. Dev Psychol. 1992;28:1030–7.
Battaglia M, Bajo S, Strambi LF, Brambilla F, Castronovo C, Vanni G, et al. Physiological and behavioral responses to minor stressors in offspring of patients with panic disorder. J Psychiatr Res. 1997;31:365–76.
Rosenbaum JF, Biederman J, Gersten M, Hirshfeld DR, Meminger SR, Herman JB, et al. Behavioral inhibition in children of parents with panic disorder and agoraphobia. A controlled study. Arch Gen Psychiatry. 1988;45:463–70.
Rosenbaum JF, Biederman J, Hirshfeld-Becker DR, Kagan J, Snidman N, Friedman D, et al. A controlled study of behavioral inhibition in children of parents with panic disorder and depression. Am J Psychiatry. 2000;157:2002–10.
Levey DF, Gelernter J, Polimanti R, Zhou H, Cheng Z, Aslan M, et al. Reproducible Genetic Risk Loci for Anxiety: results From ∼200,000 Participants in the Million Veteran Program. Am J Psychiatry. 2020;177:223–32.
Purves KL, Coleman JRI, Meier SM, Rayner C, Davis KAS, Cheesman R, et al. A major role for common genetic variation in anxiety disorders. Mol Psychiatry. 2019. 20 November 2019.https://doi.org/10.1038/s41380-019-0559-1.
Howard DM, Adams MJ, Clarke T-K, Hafferty JD, Gibson J, Shirali M, et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci. 2019;22:343–52.
Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium, Ripke S, Wray NR, Lewis CM, Hamilton SP, Weissman MM, et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry. 2013;18:497–511.
Nagel M, Jansen PR, Stringer S, Watanabe K, de Leeuw CA, Bryois J, et al. Meta-analysis of genome-wide association studies for neuroticism in 449,484 individuals identifies novel genetic loci and pathways. Nat Genet. 2018;50:920–7.
Coleman JRI, Gaspar HA, Bryois J. Bipolar Disorder Working Group of the Psychiatric Genomics Consortium, Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium, Breen G. The Genetics of the Mood Disorder Spectrum: genome-wide Association Analyses of More Than 185,000 Cases and 439,000 Controls. Biol Psychiatry. 2020;88:169–84.
Campbell CJ. Primates in Perspective. In: Campbell C. J., Fuentes A., MacKinnon K. C., Bearder S. K., & Stumpf R., editors. Int. Encycl. Primatol., Primates in Perspective. (2 ed.) Hoboken, NJ, USA. John Wiley & Sons, Oxford University Press Inc.; 2010. p. 1.
Richard AF, Goldstein SJ, Dewar RE. Weed macaques: the evolutionary implications of macaque feeding ecology. Int J Primatol. 1989;10:569.
Bakken TE, Miller JA, Ding S-L, Sunkin SM, Smith KA, Ng L, et al. A comprehensive transcriptional map of primate brain development. Nature 2016;535:367–75.
Rhesus Macaque Genome Sequencing and Analysis Consortium, Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–34.
Rogers J, Gibbs RA. Comparative primate genomics: emerging patterns of genome content and dynamics. Nat Rev Genet. 2014;15:347–59.
Wallis JD. Cross-species studies of orbitofrontal cortex and value-based decision-making. Nat Neurosci. 2011;15:13–19.
Golub MS, Hogrefe CE, Widaman KF, Capitanio JP. Iron deficiency anemia and affective response in rhesus monkey infants. Dev Psychobiol. 2009;51:47–59.
Rogers J, Raveendran M, Fawcett GL, Fox AS, Shelton SE, Oler JA, et al. CRHR1 genotypes, neural circuits and the diathesis for anxiety and depression. Mol Psychiatry. 2013;18:700–7.
Kalin NH, Fox AS, Kovner R, Riedel MK, Fekete EM, Roseboom PH, et al. Overexpressing Corticotropin-Releasing Factor in the Primate Amygdala Increases Anxious Temperament and Alters Its Neural Circuit. Biol Psychiatry. 2016;80:345–55.
Fox AS, Oler JA, Shelton SE, Nanda SA, Davidson RJ, Roseboom PH, et al. Central amygdala nucleus (Ce) gene expression linked to increased trait-like Ce metabolism and anxious temperament in young primates. Proc Natl Acad Sci USA. 2012;109:18108–13.
Fox AS, Souaiaia T, Oler JA, Kovner R, Kim JMH, Nguyen J, et al. Dorsal Amygdala Neurotrophin-3 Decreases Anxious Temperament in Primates. Biol Psychiatry. 2019;86:881–9.
Capitanio JP. Naturally Occurring Nonhuman Primate Models of Psychosocial Processes. ILAR J. 2017;58:226–34.
Bimber BN, Ramakrishnan R, Cervera-Juanes R, Madhira R, Peterson SM, Norgren RB, et al. Whole Genome Sequencing Predicts Novel Human Disease Models in Rhesus Macaques. Genomics. 2017;109:214–20.
Fawcett GL, Raveendran M, Deiros DR, Chen D, Yu F, Harris RA, et al. Characterization of single-nucleotide variation in Indian-origin rhesus macaques (Macaca mulatta). BMC Genom. 2011;12:311.
Rogers J, Garcia R, Shelledy W, Kaplan J, Arya A, Johnson Z, et al. An initial genetic linkage map of the rhesus macaque (Macaca mulatta) genome using human microsatellite loci. Genomics. 2006;87:30–8.
Xue C, Raveendran M, Harris RA, Fawcett GL, Liu X, White S, et al. The population genomics of rhesus macaques (Macaca mulatta) based on whole-genome sequences. Genome Res. 2016;26:1651–62.
Warren WC, Harris RA, Haukness M, Fiddes IT, Murali SC, Fernandes J, et al. Sequence diversity analyses of an improved rhesus macaque genome enhance its biomedical utility. Science. 2020;370:eabc6617. https://doi.org/10.1126/science.abc6617.
Glahn DC, Nimgaonkar VL, Raventós H, Contreras J, McIntosh AM, Thomson PA, et al. Rediscovering the value of families for psychiatric genetics research. Mol Psychiatry. 2019;24:523–35.
Cressie N, Read TRC. Multinomial Goodness-of-Fit Tests. J R Stat Soc Ser B Methodol. 1984;46:440–64.
Yates F. Contingency Tables Involving Small Numbers and the χ2 Test. Suppl. J R Stat Soc. 1934;1:217–35.
Seabold S, Perktold J. Statsmodels: Econometric and statistical modeling with python. Proceedings of the 9th Python in Science Conference. 2010.
Vallat R. Pingouin: statistics in Python. J Open Source Softw. 2018;3:1026.
Tromp DPM, Williams LE, Fox AS, Oler JA, Roseboom PH, Rogers GM, et al. Altered Uncinate Fasciculus Microstructure in Childhood Anxiety Disorders in Boys But Not Girls. Am J Psychiatry. 2019;176:208–16.
Fox AS, Oler JA, Shackman AJ, Shelton SE, Raveendran M, McKay DR, et al. Intergenerational neural mediators of early-life anxious temperament. Proc Natl Acad Sci USA. 2015;112:9118–22.
Oler JA, Fox AS, Shelton SE, Rogers J, Dyer TD, Davidson RJ, et al. Amygdalar and hippocampal substrates of anxious temperament differ in their heritability. Nature. 2010;466:864–8.
Fox AS, Oler JA, Birn RM, Shackman AJ, Alexander AL, Kalin NH. Functional Connectivity within the Primate Extended Amygdala Is Heritable and Associated with Early-Life Anxious Temperament. J Soc Neurosci. 2018;38:7611–21.
Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinforma. 2013;43:11.10.1–11.10.33.
McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, Thormann A, et al. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122.
Lippert C, Listgarten J, Liu Y, Kadie CM, Davidson RI, Heckerman D. FaST linear mixed models for genome-wide association studies. Nat Methods. 2011;8:833–5.
Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47:D886–94.
Kovner R, Souaiaia T, Fox AS, French DA, Goss CE, Roseboom PH, et al. Transcriptional Profiling of Primate Central Nucleus of the Amygdala Neurons to Understand the Molecular Underpinnings of Early-Life Anxious Temperament. Biol Psychiatry. 2020:S000632232031595X.
Beesdo K, Bittner A, Pine DS, Stein MB, Höfler M, Lieb R, et al. Incidence of social anxiety disorder and the consistent risk for secondary depression in the first three decades of life. Arch Gen Psychiatry. 2007;64:903–12.
Chronis-Tuscano A, Degnan KA, Pine DS, Perez-Edgar K, Henderson HA, Diaz Y, et al. Stable early maternal report of behavioral inhibition predicts lifetime social anxiety disorder in adolescence. J Am Acad Child Adolesc Psychiatry. 2009;48:928–35.
Essex MJ, Klein MH, Slattery MJ, Goldsmith HH, Kalin NH. Early risk factors and developmental pathways to chronic high inhibition and social anxiety disorder in adolescence. Am J Psychiatry. 2010;167:40–6.
Hirshfeld-Becker DR, Biederman J, Henin A, Faraone SV, Davis S, Harrington K, et al. Behavioral inhibition in preschool children at risk is a specific predictor of middle childhood social anxiety: a five-year follow-up. J Dev Behav Pediatr JDBP. 2007;28:225–33.
Williamson DE, Coleman K, Bacanu S-A, Devlin BJ, Rogers J, Ryan ND, et al. Heritability of fearful-anxious endophenotypes in infant rhesus macaques: a preliminary report. Biol Psychiatry. 2003;53:284–91.
Coyne SP, Lindell SG, Clemente J, Barr CS, Parker KJ, Maestripieri D. Dopamine D4 receptor genotype variation in free-ranging rhesus macaques and its association with juvenile behavior. Behav Brain Res. 2015;292:50–5.
Kinnally EL, Karere GM, Lyons LA, Mendoza SP, Mason WA, Capitanio JP. Serotonin pathway gene-gene and gene-environment interactions influence behavioral stress response in infant rhesus macaques. Dev Psychopathol. 2010;22:35–44.
Bergen SE, Gardner CO, Kendler KS. Age-related changes in heritability of behavioral phenotypes over adolescence and young adulthood: a meta-analysis. Twin Res Hum Genet J Int Soc Twin Stud. 2007;10:423–33.
Waszczuk MA, Zavos HMS, Gregory AM, Eley TC. The phenotypic and genetic structure of depression and anxiety disorder symptoms in childhood, adolescence, and young adulthood. JAMA Psychiatry. 2014;71:905–16.
Park C, Falls W, Finger JH, Longo-Guess CM, Ackerman SL. Deletion in Catna2, encoding αN-catenin, causes cerebellar and hippocampal lamination defects and impaired startle modulation. Nat Genet. 2002;31:279–84.
Ehlers CL, Gizer IR, Bizon C, Slutske W, Peng Q, Schork NJ, et al. Single Nucleotide Polymorphisms in the REG-CTNNA2 region of chromosome 2 and NEIL3 associated with impulsivity in a Native American sample. Genes Brain Behav. 2016;15:568–77.
Terracciano A, Esko T, Sutin AR, de Moor MHM, Meirelles O, Zhu G, et al. Meta-analysis of genome-wide association studies identifies common variants in CTNNA2 associated with excitement-seeking. Transl Psychiatry. 2011;1:e49.
Edwards AC, Aliev F, Bierut LJ, Bucholz KK, Edenberg H, Hesselbrock V, et al. Genome-wide association study of comorbid depressive syndrome and alcohol dependence. Psychiatr Genet. 2012;22:31–41.
Sun Y, Chang S, Liu Z, Zhang L, Wang F, Yue W, et al. Identification of novel risk loci with shared effects on alcoholism, heroin, and methamphetamine dependence. Mol Psychiatry. 2019;26;1152–61.
Boraska V, Franklin CS, Floyd JAB, Thornton LM, Huckins LM, Southam L. et al. A genome-wide association study of anorexia nervosa. Mol Psychiatry. 2014;19:1085–94.
Johnston KJA, Adams MJ, Nicholl BI, Ward J, Strawbridge RJ, Ferguson A, et al. Genome-wide association study of multisite chronic pain in UK Biobank. PLoS Genet. 2019;15:e1008164.
Schaffer AE, Breuss MW, Caglayan AO, Al-Sanaa N, Al-Abdulwahed HY, Kaymakçalan H, et al. Biallelic loss of human CTNNA2, encoding αN-catenin, leads to ARP2/3 complex overactivity and disordered cortical neuronal migration. Nat Genet. 2018;50:1093–101.
Acknowledgements
We would like to thank the staff at the California National Primate Research Center, Harsha Doddapaneni, Donna Muzny and the HGSC data production team, Dr William Mason for helping to develop the Food Retrieval Task, as well as H. Sompura. This work was supported by NIH grants OD010962 to JPC, R01MH121735 to ASF, OD011173 to JR, and the California National Primate Research Center (P51OD011107).
Author information
Authors and Affiliations
Contributions
FAS: Conceptualization, Formal Analysis, Methodology, Software, Visualization, Writing—original draft (lead); HRA: Data curation, Formal Analysis, Software, Visualization, Writing—review & editing; LDR: Conceptualization (Food Retrieval), Investigation (BBA and Food Retrieval) (lead), Project Administration (BBA program) (lead), Supervision (BBA) (lead), Writing—review and editing; MR: Data curation, Formal analysis, Project administration; KS: Writing—original draft; ELK: Resources (provided DNA), Writing—review and editing. CJP: Conceptualization (BBA program), Data curation (BBA), Methodology (BBA), Resources (BBA data), Writing—review and editing; RJ: Conceptualization, Supervision, Writing—review and editing.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Fox, A.S., Harris, R.A., Rosso, L.D. et al. Infant inhibited temperament in primates predicts adult behavior, is heritable, and is associated with anxiety-relevant genetic variation. Mol Psychiatry 26, 6609–6618 (2021). https://doi.org/10.1038/s41380-021-01156-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-021-01156-4
This article is cited by
-
Rhesus infant nervous temperament predicts peri-adolescent central amygdala metabolism & behavioral inhibition measured by a machine-learning approach
Translational Psychiatry (2024)
-
Nonhuman primate genetic models for the study of rare diseases
Orphanet Journal of Rare Diseases (2023)
-
Current advances in primate genomics: novel approaches for understanding evolution and disease
Nature Reviews Genetics (2023)