Abstract
Leptin is an adipocyte-derived hormone with pleiotropic functions affecting appetite and mood. While leptin’s role in the regulation of appetite has been extensively studied in hypothalamic neurons, its function in the hippocampus, where it regulates mood-related behaviors, is poorly understood. Here, we show that the leptin receptor (LepRb) colocalizes with brain-derived neurotrophic factor (BDNF), a key player in the pathophysiology of major depression and the action of antidepressants, in the dentate gyrus of the hippocampus. Leptin treatment increases, whereas deficiency of leptin or leptin receptors decreases, total Bdnf mRNA levels, with distinct expression profiles of specific exons, in the hippocampus. Epigenetic analyses reveal that histone modifications, but not DNA methylation, underlie exon-specific transcription of the Bdnf gene induced by leptin. This is mediated by stimulation of AKT signaling, which in turn activates histone acetyltransferase p300 (p300 HAT), leading to changes in histone H3 acetylation and methylation at specific Bdnf promoters. Furthermore, deletion of Bdnf in the dentate gyrus, or specifically in LepRb-expressing neurons, abolishes the antidepressant-like effects of leptin. These findings indicate that leptin, acting via an AKT-p300 HAT epigenetic cascade, induces exon-specific Bdnf expression, which in turn is indispensable for leptin-induced antidepressant-like effects.
Similar content being viewed by others
Introduction
Leptin is produced and secreted from adipocytes [1], circulates in the blood [2], and is transported across the blood–brain barrier [3]. Leptin’s target neurons are distributed widely throughout the brain [4,5,6], through which leptin exerts pleiotropic effects by activating the signaling form of the leptin receptor (LepRb) [7,8,9]. While extensive studies have focused on leptin’s actions in the hypothalamus in the regulation of feeding and energy metabolism [10, 11], LepRb is also highly expressed in extra-hypothalamic brain regions including the hippocampus, which is implicated in mood regulation [4, 12,13,14]. We and others have shown that circulating leptin levels are reduced in chronic stress animal models of depression [15,16,17], whereas systemic and intracerebroventricular injections of leptin produce antidepressant-like behavioral effects [15, 18,19,20]. In humans, leptin levels were found to correlate negatively with the severity of depression symptoms [21], Multiple lines of evidence suggest that leptin targets hippocampal neurons to regulate depression-related behaviors. First, direct infusion of leptin into the dentate gyrus of the hippocampus induces an antidepressant-like effect, similar to the effect observed after systemic injection [15]. Second, deletion of LepRb in the dentate gyrus results in depression-like behaviors and attenuates leptin’s antidepressant-like effects [13, 22]. Third, blockade of leptin signaling in the dentate gyrus attenuates the antidepressant-like effects of leptin [14]. These findings support an important role of the hippocampus in mediating leptin’s actions on mood-related behavior. However, the precise molecular mechanisms remain unknown.
Brain-derived neurotrophic factor (BDNF) is a neurotrophin that has been implicated in the pathophysiology of depression and the mechanisms of action of antidepressant drugs [23]. Hippocampal mRNA or protein levels of BDNF are increased by treatment with different classes of antidepressants [24, 25] in rodents and in depressed patients [26]. Antidepressant drugs activate the BDNF receptor, tropomycin receptor kinase B (TrkB), in the hippocampus, suggesting enhanced BDNF release [27]. Furthermore, infusion of BDNF directly into the hippocampus is sufficient to induce antidepressant-like effects [28, 29]. Importantly, the antidepressant-like response to classical antidepressants is attenuated in transgenic mice with disrupted BDNF signaling [27]. In addition, the rapid onset antidepressant effects of ketamine require expression of BDNF [30, 31]. Together, these data indicate that increased BDNF expression and signaling are both necessary and sufficient to produce the behavioral effects of antidepressants.
The Bdnf gene in rodents contains eight noncoding 5′ exons (I–VIII), which are alternatively spliced to the 3′ protein-encoding exon IX [32, 33]. Each exon is driven by its own unique promoter sequence for the temporal and spatial specificity of Bdnf expression in an activity-dependent manner [34, 35]. All Bdnf mRNA isoforms are translated into the same BDNF protein [36]. Specific exon-containing Bdnf mRNAs are differentially regulated by stimuli such as changes in neuronal activity [37,38,39], stress [40,41,42,43], and antidepressant treatment [44,45,46,47]. However, whether leptin regulates the transcription of the Bdnf gene in the hippocampus is unknown.
In this study, we investigated (1) whether, and to what extent, LepRb is colocalized with BDNF in the hippocampus; (2) whether and how leptin regulates exon-specific Bdnf expression; (3) whether leptin and deficiencies of leptin or its receptor have opposite effects; and (4) whether Bdnf expression in the hippocampus is required for leptin’s antidepressant-like behavioral effects. To address these questions, we employed a genetic approach to investigate colocalization of LepRb and BDNF expression in the brain. Furthermore, we examined exon-specific Bdnf expression in response to leptin treatment and impaired leptin signaling and further explored the epigenetic mechanisms underlying leptin-induced regulation of Bdnf gene expression. Finally, we determined the effects of loss of BDNF in the hippocampus on behavioral responses to leptin.
Materials and methods
Animals
Wild-type C57BL/6J mice (Stock No. 000664), LepRb-ires-Cre mice (Stock No. 008320), Ai14-tdTomato mice (Stock No. 007914), Bdnfflox/flox (Stock No. 004339), Bdnfklox/klox mice (Stock No. 021055), and Emx1-Cre mice (Stock No. 005628) were purchased from Jackson Laboratory (Bar Harbor, ME, USA) and maintained as breeding colonies. The ob/ob and db/db mice and their littermates were obtained by intercrossing ob/+ or db/+ mice (ob/+, Stock No. 000632; db/+, Stock No. 000642; Jackson Laboratory). LepRb-ires-Cre mice were generated with an IRES-NLS-Cre cassette “knocked in” to the region immediately 3′ to the LepRb stop codon [48] to drive Cre expression in all Lepr-expressing cells. Ai14 mice have a loxP-flanked STOP cassette preventing transcription of a CAG promoter-driven tdTomato protein in all cells. Ai14 mice express robust tdTomato fluorescence following Cre-mediated recombination [49]. Bdnfflox/flox mice possess loxP sites on either side of the Bdnf coding region [50]. Bdnfklox/klox mice possess loxP sites flanking the Bdnf encoding exon 9, with a polyadenylation sequence upstream of the 3′ loxP site, and lacZ downstream of the 3′ loxP site [51]. All mice were maintained in the C57BL/6J background. Mice were housed in a group of 5 and maintained on a 12 h light/12 h dark cycle with ad libitum access to food and water. The animal protocols used in this study were approved by the Institutional Animal Care and Use Committees of the University of Texas Health Science Center at San Antonio, Binzhou Medical University Hospital, and Augusta University.
To label LepRb neurons, LepRb-ires-Cre mice were crossed with the reporter Ai14-tdTomato mice to produce LepRb-tdTomato reporter mice [14]. To examine the colocation of LepRb and BDNF, LepRb-ires-Cre mice were crossed with Bdnfklox/klox mice. The Bdnfflox/+;Lepr-ires-Cre offspring were used to detect the presence of β-galactosidase encoded by lacZ. To generate conditional knockout mice lacking Bdnf in Lepr neurons, Bdnfflox/flox mice were crossed with LepRb-ires-Cre mice. The Bdnfflox/+;Lepr-ires-Cre offspring were back-crossed with Bdnfflox/flox to produce Bdnfflox/flox;Lepr-ires-Cre and Bdnfflox/flox littermates for the experiments. Leprflox/flox;Emx1-Cre mice were generated as previously described [13]. Briefly, Emx1-Cre mice were crossed with Leprflox/flox mice. The Leprflox/+;Emx1-Creoffspring were subsequently crossed with Leprflox/flox mice to generate Leprflox/flox;Emx1-Cre mice and Leprflox/flox littermate controls. The PCR primers used for genotyping were as follows: LepRb-ires-Cre: forward-5′-GCGGTCTGGCAGTAAAAACTATC-3′, reverse-5′-GTG AAACAGCATTGCTGTCACTT-3′; Bdnfflox/flox: forward-5′-TGTGATTGTGTTTCTGGTGAC-3′, reverse-5′-GCCTTCATGCAACC GAAGTATG-3′; Bdnfklox/klox: forward-5′-CTTGGGTGGAGAGGCTATTC-3′, reverse-5′-AGGTGAGATGACAGGAGATC-3′; ob/ob: primer1: 5-GCAGTCGGTATCCGCCAAGCAG-3′, primer2: -5′-GTGGTCTACAGGAGGGAGAGAAATG-3′; SNP1: 5′-TAGCCAATGACCTGGAGA ATCACT-3′; SNP2: 5′-CCAGCAGATGGAGGAGGTCACG-3′; and db/db: primer1: 5′-TTTTTATTTTGCTTGCTTATTTTGT-3′, primer2: 5′-GATTTGAACTCAGGACCTTTGG-3′; SNP1:5′-GATGTTTACATTTTGATGGACGT-3′; SNP2: 5′-TCAAACCATAGTTTAGGTTTGTATC-3′; and Emx1-Cre: forward-5′-CACTCATGGAAAATAGCGATC-3′, reverse-5′-ATCTCCGGTATTGAAACTCCAGCGC-3′.
Drugs
Recombinant mouse leptin (R&D Systems, Minneapolis, MN, USA) was dissolved in sterile saline and administered intraperitoneally (i.p.) at a dose of 1 or 5 mg/kg body weight. For intra-dentate gyrus injections, AKT inhibitor (AKTi) VIII (1,3-dihydro-1-(1-((4-(6-Phenyl-1H-imidazo[4,5-g]quinoxalin-7-yl)phenyl)methyl)-4-piperidinyl)-2H-benzimidazol-2-one), an isozyme-selective AKTi specific for AKT1/2 (Calbiochem, La Jolla, CA, USA), was dissolved in dimethylsulfoxide (Sigma-Aldrich, Saint Louis, MO, USA) and injected at a dose of 3.0 μg/μl. C646, a specific inhibitor of histone acetyltransferase p300 (p300 HAT; Sigma-Aldrich) was dissolved in dimethylsulfoxide and injected at a dose of 3 μg/μl.
Immunohistochemistry
Lepr-ires-cretdTomato and Bdnfklox/+;Lepr-ires-cre mice were transcardially perfused with 4% paraformaldehyde. The brains were removed, postfixed overnight and then cryoprotected in 30% sucrose and cut into 20-μm coronal sections. From serial coronal sections of the entire dorsal-ventral axis of dentate gyrus (from −1.34 to −3.52 mm posterior to bregma), every sixth section was selected from each animal and processed for immunohistochemical staining. The sections were rinsed three times in phosphate-buffered saline (PBS), and incubated in blocking buffer (1% bovine serum albumin, 3% goat serum, 0.3% Triton X-100 in PBS) for 1 h. The sections were then incubated with mouse anti-NeuN antibody (#ab104224, 1:500; Abcam, Cambridge, UK) and rabbit anti-β-galactosidase antibody (#ab4761, 1:1000, Abcam, Cambridge, UK) overnight at 4 °C. After washing in PBS, sections were incubated for 4 h with fluorescent secondary antibodies: Alexa Fluor® 488 donkey antimouse IgG (#A-21202, 1:400, Invitrogen, Carlsbad, CA, USA) and Alexa Fluor® 555 donkey antirabbit IgG (#A-31572, 1:400, Invitrogen, Carlsbad, CA, USA). Finally, the sections were washed in PBS, mounted onto poly-lysine-coated glass slides, cover-slipped using fluorescence mounting medium and visualized via an Olympus FV1000 confocal microscope (Olympus, Shinjuku, Tokyo, Japan). Cell numbers were quantified on every sixth section throughout the dorsal-ventral axis of hippocampus using unbiased stereology. LepRb-tdTomato-, β-galactosidase-, and NeuN-positive cells within both sides of the hippocampus were counted. Fluorescent cells that intersected the exclusion boundaries of the unbiased sampling frame were excluded from counting. Cells that met the counting criteria were counted bilaterally throughout the dentate gyrus. The colocalization of LepRb-tdTomato or β-galactosidase with NeuN was confirmed with z-stack of the respective cell soma using line-based sequential scan (4-μm interval). The percentage of NeuN-positive neurons that were also double-labeled for LepRb-tdTomato and β-galactosidase within the granule cell layer was calculated.
Western blot assay
Mice were killed by rapid decapitation. The hippocampus was dissected out on ice and immediately homogenized in the lysis buffer with 1% phenylmethylsulfonyl fluoride and 1 × PhosSTOP phosphatase inhibitor cocktail (Roche Applied Science, Penzberg, Germany). The denatured protein was separated on an SDS-polyacrylamide gel electrophoresis and transferred to a polyvinylidene fluoride membrane. The membrane was blocked in a solution of Tris-buffered saline with 1% dried milk and 0.1% Tween 20. Membranes were then incubated with the following primary antibodies diluted in the blocking solution: anti-AKT (#9272, 1:1000, Cell Signaling Technology, Danvers, MA, USA), anti-phospho-AKT (Thr308) (#2965, 1:1000, Cell Signaling), anti-BDNF (sc-546, 1:500, Santa Cruz Biotechnology), anti-ERK1/2 (#9102, 1:1000, Cell Signaling), anti-phospho-ERK1/2 (Thr202/Tyr204) (#4370, 1:1000, Cell Signaling), anti-TrkB (#4603, 1:1000, Cell Signaling), anti-pTrkB (Tyr516) (#4619, 1:1000, Cell Signaling), anti-β-actin (#4970, 1:1000, Cell Signaling), anti-STAT3 (#9139, 1:1000, Cell Signaling), anti-p-STAT3 (#9145, 1:1000, Cell Signaling). After washing, membranes were incubated with goat antirabbit IR Dye 680LT (#926-68021, 1:5000, Li-COR Biosciences, Lincoln, NE, USA) or goat antimouse IR Dye 800CW (#926-32210, 1:5000, Li-COR Biosciences) fluorescent secondary antibodies secondary antibodies and visualized using an Odyssey infrared imaging system (Li-COR Biosciences).
Reverse transcription (RT) and quantitative PCR
The hippocampus was isolated from the mouse brain as described above and dentate gyrus tissue was dissected from 100-μm-thick coronal brain slices (bregma: −1.34 mm to −3.52 mm). Total RNA was extracted from brain tissue to generate cDNA using a previously described protocol [52]. First, total RNA was treated with 4 × gDNA wiper mix at 42 °C for 2 min to remove genomic DNA contamination, then reverse transcribed into cDNAs with 5 × HiScript II QRT SuperMix that was added to the reaction mixture and incubated at 25 °C for 10 min followed by 50 °C for 30 min and 85 °C for 5 min. The resulting cDNA was used for real-time PCR detection using the StepOnePlus real-time PCR system (Applied Biosystems, Waltham, MA, USA). The condition for PCR was 95 °C for 5 min, followed by 40 cycles of 95 °C for 10 s and 60 °C for 30 s. The primer sequences used to amplify each desired product were as follows: Bdnf exon IX (forward: GCGCCCATGAAAGAAGTAAA; reverse: TCGTCAGACCTCTCGAACCT), exon I (forward: 5′-CCTGCATCTGTTGGG GAGAC-3′; reverse: 5′-GCCTTGTCCGTGGACGTTTA-3′), exon II (forward: 5′-CTAGCCACCGGGGTGGTGTAA-3′; reverse: 5′-AGGATGGTCATCACTCTTCTC-3′), exon III (forward: 5′-CTTCATTGAGCCCAGGTCC-3′; reverse: 5′-CCGTGGACGTTTACTTCTTTC-3′), exon IV (forward: 5′-CAGAGCAGCTGCCTTGATGTT-3′; reverse: 5′-GCCTTGTCCGTGGACGTTTA-3′), exon VI (forward: 5′-CTGGGAGGCTTTGATGAGAC-3′; reverse: 5′-GCCTTCATGCAACCGAAGTA-3′), Cre (forward: 5′-GATTTCGACCAGGTTCGTTC-3′; reverse: 5′- GCTAACCAGCGTTTTCGTTC-3′), and β-tubulin (forward: 5′-AGCAACATGAATGACCTGGTG-3′; reverse: 5′-GCTTTCCCTAACCTGCTTGG-3′). The housekeeping gene β-tubulin was used as a reference gene for normalization of gene expression. The 2−ΔΔCT method, i.e., delta-delta-ct analysis, was used as a relative quantification.
Chromatin immunoprecipitation (ChIP) analysis
The hippocampus was dissected out from the brain on ice and incubated in 1% formaldehyde in PBS for 10 min followed by incubation with 125 mM glycine for 10 min at room temperature. Tissues were then washed three times in PBS and homogenized in SDS lysis buffer (167 mM NaCl, 0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.1) containing PhosSTOP Phosphatase Inhibitor Cocktail (Roche Applied Science, Penzberg, Germany). Tissue samples were sonicated on ice using a sonicator (FB-705; Fisher Scientific) at 50% amplitude, 5 s on/10 s off, for 5 min. Samples were then centrifuged at 12,000 rpm for 10 min at 4 °C, and supernatants were collected for dilution in the ChIP dilution buffer (167 mM NaCl, 0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.1). Then, the resulting chromatin solution was precleared with 60 µ salmon sperm DNA/protein A-agarose (EMD Millipore, Burlington, MA, USA) before binding with mouse IgG (#12-371, Sigma-Aldrich) or rabbit IgG (#12-370, Sigma-Aldrich) for 3–4 h at 4 °C and centrifuged for 2 min at 3000 rpm. Immunoprecipitations were performed at 4 °C overnight with primary antibodies: antiacetyl H3 (#06–599, EMD Millipore), antiacetyl H4 (#06-866, Millipore), anti-H3K4me2 (#ab7766, Abcam), anti-H3K9me2 (#ab1220, Abcam), anti-p300 HAT (#05–257, EMD Millipore), anti-H3K27me2 (#ab24684, Abcam). DNA-histone complex was collected by binding to salmon sperm DNA blocked-protein A-agarose beads for a 3–4 h incubation at 4 °C and sequentially washed with low salt buffer (150 mM NaCl, 0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1), high salt buffer (500 mM NaCl, 0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1), LiCl immune complex buffer (0.25 M LiCl, 1% IGEPAL-CA630, 1% deoxycholic acid, 1 mM EDTA, 10 mM Tris, pH 8.1) and Tris-EDTA buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0). DNA-histone complexes were removed from protein A-agarose beads by incubation with elution buffer (0.1 M NaHCO3, 1% SDS) for 15 min at room temperature followed by incubation with NaCl (final concentration 0.2 M) overnight at 65 °C to dissociate DNA and histones. After proteinase K digestion for 2 h at 45 °C, DNA was extracted using DNA extraction Kits (Omega bio-tek, Doraville, GA, USA). Immunoprecipitated DNA was subjected to quantitative real-time PCR using primers specific to the mouse Bdnf promoters or GAPDH: Bdnf promoter I: forward-5′-TGATCATCACTCACGACCACG-3′, Reverse-5′- CAGCCTCTCTGAGCCAGTTACG-3′; Bdnf promoter II: forward-5′-CCGTCTTGTATTCCATCCTTTG-3′, Reverse-5′-CCCAACTCCACCACTATCCTC-3′; Bdnf promoter III: forward-5′-GTGAGAACCTGGGGCAAATC-3′, Reverse-5′-ACGGAAAAGAGGGAGGGAAA-3′; Bdnf promoter IV: forward-5′-CTTCTGTGTGCGTGAATTTGCT-3′, Reverse-5′-AGTCCACGAGAGGGCTCCA-3′; Bdnf promoter VI: forward-5′-ACTCACACTCGCTTCCTCCT-3′, Reverse-5′-GCACTGGCTTCTCTCCATTT-3′; GAPDH: forward-5′-CTCCCAGGAAGACCCTGCTT-3′, Reverse-5′-GGAACAGGGAGGAGCAGAGA-3′.The GAPDH promoter served as an internal gene control.
DNA methylation analysis
DNA was extracted from hippocampal tissue and each sample (300 ng) was processed for bisulfite conversion using the EZ DNA Methylation Kit (Zymo Research, Orange, CA, USA). The bisulfite-converted samples were amplified using specific primers for the CpG islands that were selected from the promoter and exonic regions of the Bdnf gene for methylation analysis. The primers used to amplify six-specific CpG island regions were as follows: CpG1: forward-5′-GGGAAGTATTTAAAATAGGGTAG-3′, Reverse-5′-GGGAAGTATTTAAAATAGGGTAG-3′; CpG2: forward-5′-TTATTTAGTATTTTGGATAGAGTTAG-3′, Reverse-5′- CAATAAAAAAACCAAACTAAAACTC-3′, CpG3: forward-5′- TTTATAAAGTATGTAATGTTTTGGAA-3′, Reverse -5′- TACTCCTATTCTACAACAAAAAAATTAAAT-3′; CpG4: forward-5′- GGTATAGAGTTTTGGGTTTAAGTAG-3′, Reverse -5′- AAAATCAAACATTATTTAACTCTTC-3′; CpG5: forward-5′- TAGTGTTTGGTTTTGGTTGAGTTT-3′, Reverse -5′- CTACCCCAAAACAATAATAACAATTAAA-3′; and CpG6: forward-5′- ATTGTTATTATTGTTTTGGGGTAGA-3′, Reverse -5′- AAATTCTACAATCCCACAACTTCTC-3′. The thermocycler protocol involved an initial denaturation cycle (5 min, 95 °C), 40 cycles of denaturation (45 s, 95 °C), annealing (30 s, 56–58 °C), and extension (30 s, 72 °C), followed by a final extension cycle (5 min, 72 °C) terminating at 4 °C. The PCR products were then purified using a gel extraction kit (Qiagen, Germantown, MD, USA). The methylated CpG sites were detected using a PyroMark Q96 MD Pyrosequencing System (Qiagen) at the University of Texas Health Science Center at San Antonio Core Facility. The percentage of methylation of each CpG site within the region amplified was calculated by the MultiExperiment Viewer.
Intra-dentate gyrus microinjection
For the deletion of the Bdnf gene selectively in the dentate gyrus, anesthetized adult Bdnfflox/flox mice underwent bilateral stereotaxic injections of AAV-Cre-GFP or AAV-GFP (0.5 μl /side; Vector Biolabs, Malvern, PA) into the dentate gyrus (coordinates: AP = −2.1 mm, ML = ±1.5 mm, DV = −2.3 mm from Bregma), at a rate of 0.1 μL/min with a 33-gauge stainless steel injector connected to a UMP3 micro syringe pump (World Precision Instruments, Sarasota, FL). Additional 5 min were allowed for diffusion and prevention of backflow. Behavioral experiments were conducted 21 days after AAV injection. For intra-dentate gyrus microinjection of inhibitors of AKT and p300 HAT, the mice were anesthetized with 4% chloral hydrate (400 mg/kg, i.p.). AKTi and C646 (0.5 μl/side) were injected bilaterally into the dentate gyrus of adult C57BL/6J mice at a rate of 0.5 μl/min with a 33-gauge stainless steel injector. Additional 5 min were allowed for diffusion and prevention of backflow, and 30 min later, mice were injected with leptin (5 mg/kg, i.p.).
Estrous cycle
The stages of the estrous cycle were monitored by analysis of cell types in vaginal lavages. Vaginal lavages were collected daily between 7:00 and 8:00 a.m. as described previously [53]. Briefly, vaginal fluid was placed on slides, and the slides were stained with crystal violet. The types of cells in vaginal smears were examined under a light microscope. Diestrus was defined by the presence of small leukocytes, and proestrus was defined by the presence of clumps of large, round, nucleated epithelial cells.
Forced swim test
Mice were placed into a clear Plexiglas cylinder (25 cm in height and 10 cm in diameter) filled with water (24 °C) to a depth of 15 cm. A 6-min swim session was videotaped by a camera mounted above the cylinder. The duration of immobility was measured for the last 4 min. Immobility was defined as the absence of all movements except those required for respiration.
Tail suspension test
The apparatus consisted of a box (30 × 30 × 30 cm) with an open front and a bar placed horizontally 1 cm from the top with an attached vertical bar hanging down in the center. Mice were individually suspended by the tail on the vertical bar with adhesive tape affixed 2 cm from the tip of the tail. A camera positioned in front of the box was used to record the animals’ behavior for a 6-min test session. Immobility in this test was defined as the absence of any limb or body movements, except those caused by respiration.
The behaviors of each mouse were scored by experimenters who were blinded to the genotypes or treatment conditions.
Statistical analysis
All statistical analyses were performed using the statistical software GraphPad Prism 8. Shapiro–Wilk test and F-test were used to test normality and equal variance assumptions, respectively. For normally distributed data, two-tailed t-tests were used to assess differences between two experimental groups with equal variance. For normally distributed data with unequal variances, two-tailed t-tests with Welch’s correction were used. One-way analyses of variance (ANOVAs) followed by Sidak post hoc tests were used for analysis of three or more groups. For nonnormally distributed data, Mann–Whitney U tests were performed to compare two groups. For analysis of three or more groups with nonnormally distributed data, the Kruskal–Wallis test followed by Dunn’s multiple comparisons test was used. Two-way or three-way ANOVAs followed by Bonferroni tests were used where appropriate. P < 0.05 was considered significant.
Results
Colocalization of LepRb and BDNF in the dentate gyrus of the hippocampus
While alternative splicing produces multiple isoforms of the LepR, the long isoform, LepRb, has an intracellular domain that is essential for signal transduction required to elicit the physiological effects of leptin [54, 55]. We examined the distribution of LepRb-expressing neurons in the brain using LepRb-ires-Cre × Ai14-tdTomato (LepRb-tdTomato) reporter mice, in which tdTomato marks LepRb-expressing cells [4, 14]. As shown previously [4, 14, 56], leptin target neurons were found in the hypothalamus, prefrontal cortex, hippocampus, and ventral tegmental area (Fig. 1a). To examine if these neurons express BDNF, we crossed LepR-ires-Cre mice with Bdnfklox/+ mice, in which BDNF neurons express β-galactosidase once the floxed Bdnf allele is deleted by Cre-mediated recombination [51, 57]. In Bdnfklox/+;LepR-ires-Cre mice, very few β-galactosidase-expressing neurons were detected in the hypothalamus, prefrontal cortex, or ventral tegmental area, indicating lack of co-expression between LepRb and BDNF in these brain regions. However, dense populations of granule neurons in the dentate gyrus of the hippocampus expressed β-galactosidase (Fig. 1a), suggesting the co-expression of LepRb and BDNF.
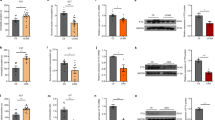
a Colocalization of LepRb and BDNF. Upper panel: schematic diagram of the strategy used for generation of tdTomatoLepr-ires-Cre (left) and Bdnfklox/+;LepR-ires-Cre mice (right). Lower panel: representative confocal images showing LepRb-expressing neurons labeled with tdTomato (red) in LepRb-CretdTomato mice and neurons coexpressing LepRb and BDNF labeled with β-galactosidase (green) in Bdnfklox/+;LepR-ires-Cre mice. Sale bars = 100 μm. DG dentate gyrus; GCL granule cell layer; PFC prefrontal cortex; VTA ventral tegmental area. b Distribution of LepRb-expressing neurons in the dentate gyrus. Left, representative images showing LepRb-expressing neurons (red) and NeuN-positive cells (green) in the dentate gyrus. Right, quantitative analysis of dentate gyrus neurons expressing LepRb in the suprapyramidal versus infrapyramidal blade and in the dorsal versus ventral region. n = 4 mice. c Distribution of neurons coexpressing LepRb and BDNF in the dentate gyrus. Left, representative images showing β-galactosidase-positive cells, indicative of LepRb/BDNF-coexpressing neurons (red), and NeuN-positive cells (green). Right, quantitative analysis of dentate gyrus neurons coexpressing LepRb and BDNF in the subcompartments of the dentate gyrus. n = 4 mice. Sale bars = 100 μm for low magnification images and 20 μm for high magnification images. *P < 0.05; **P < 0.01; ***P < 0.001.
The dentate gyrus is composed of the suprapyramidal and infrapyramidal blades, and the dorsal and ventral regions are functionally distinct [58]. Therefore, we analyzed the distribution of neurons expressing LepRb alone and coexpressing LepRb/BDNF in these subcompartments of the dentate gyrus. In LepRb-tdTomato reporter mice, LepRb neurons were labeled with tdTomato. We quantified the numbers of LepRb neurons and total NeuN-positive cells in the suprapyramidal versus infrapyramidal blade and in the dorsal versus ventral region. Data analysis revealed that a higher proportion of neurons in the ventral region expressed LepRb (blade: F(1,12) = 1.8970, P = 0.1936; region: F(1,12) = 28.4700, P < 0.001; total number of LepRb neurons counted, 2380; total number of NeuN-positive neurons counted, 27,155 from 4 mice) (Fig. 1b). Furthermore, we quantified the numbers of β-galactosidase-positive cells, indicative of colocalization of LepRb and BDNF, and total NeuN-positive cells in Bdnfklox/+;LepR-ires-Cre mice. Cell counting results indicated a region-specific distribution with a higher percentage of neurons in the dorsal region expressing both LepRb and BDNF (blade: F(1,12) = 0.1587, P = 0.6973; region: F(1,12) = 144.5000, P < 0.001; total number of LepRb/BDNF neurons counted, 2029; total number of NeuN-positive neurons counted, 37,318 from 4 mice) (Fig. 1c). By comparing the numbers and the distribution of LepRb-expressing neurons with LepRb/BDNF-colocalized neurons in the dentate gyrus, we determined that a high proportion of LepRb-expressing neurons (up to 79%) contain detectable BDNF especially in the dorsal region, which provide an anatomical basis to study functional interactions between LepRb signaling and BDNF.
Leptin regulates BDNF mRNA and protein expression in the hippocampus
We next examined whether leptin regulates Bdnf mRNA and protein expression. Specifically, we measured Bdnf exon IX mRNA levels using quantitative real-time PCR. The detection of exon IX allows for the evaluation of all isoforms transcribed, i.e., total Bdnf transcription. Wild-type mice received a single i.p. injection of leptin (1 or 5 mg/kg), and total Bdnf mRNA levels in the brain were measured 30 min or 2 h post injection. Leptin treatment caused no significant change in total Bdnf mRNA expression in the hippocampus at 30 min post injection (1 mg/kg, t(11) = 1.4330, P = 0.1797; 5 mg/kg, t-test with Welch’s correction, P = 0.8218) (Fig. 2a); however, an increase was observed at 2 h post injection (1 mg/kg, t-test with Welch’s correction, P = 0.0272; 5 mg/kg, Mann–Whitney test, P = 0.0011) (Fig. 2a). In contrast, total Bdnf mRNA levels in the PFC and hypothalamus remained unaltered 2 h following the same leptin treatments (PFC, 1 mg/kg, Mann–Whitney test, P = 0.1255; 5 mg/kg, Mann–Whitney test, P = 0.8328; hypothalamus, 1 mg/kg, Mann–Whitney test, P = 0.1775; 5 mg/kg, Mann–Whitney test, P = 0.6282) (Fig. 2a). In parallel, BDNF protein levels in the hippocampus were increased 2 h after leptin treatment (1 or 5 mg/kg) (1 mg/kg, t(10) = 2.2810, P = 0.0457; 5 mg/kg, t(11) = 2.5670, P = 0.0262) (Fig. 2b). The increase in BDNF protein levels persisted at least 6 h after an i.p. injection of 5 mg/kg of leptin (t(8) = 2.8310, P = 0.0221). However, this was not observed after an i.p. injection of a lower dose of leptin (1 mg/kg) (t(10) = 0.0470, P = 0.9634) (Fig. 2b).
a Total (exon IX) Bdnf mRNA levels in the hippocampus, prefrontal cortex (PFC), and hypothalamus at 30 min or 2 h after leptin injection (1 mg/kg or 5 mg/kg, i.p.). Hippocampus-30 min: saline, n = 7; leptin (1 mg/kg), n = 6; saline, n = 7; leptin (5 mg/kg), n = 6. Hippocampus-2 h: saline, n = 12; leptin (1 mg/kg), n = 11; saline, n = 10; leptin (5 mg/kg), n = 11. PFC-2 h: saline, n = 6; leptin (1 mg/kg), n = 5; saline, n = 11; leptin (5 mg/kg), n = 12. Hypothalamus-2 h: saline, n = 6; leptin (1 mg/kg), n = 5; saline, n = 6, leptin (5 mg/kg), n = 7. b BDNF protein levels in hippocampus in response to leptin treatment. Post-injection 2 h: saline, n = 6; leptin (1 mg/kg), n = 6; saline, n = 6, leptin (5 mg/kg), n = 7. Post-injection 6 h: saline, n = 6; leptin (1 mg/kg), n = 6; saline, n = 5; leptin (5 mg/kg), n = 5. c Phosphorylated TrkB levels in the hippocampus at 2 and 6 h after leptin injection (5 mg/kg). Post-injection 2 h: saline, n = 6; leptin, n = 5. Post-injection 6 h: saline, n = 5; leptin, n = 5. *P < 0.05; **P < 0.01 compared with saline-injected controls.
Most biological actions of BDNF are transduced through TrkB receptors [27, 59, 60]. Therefore, we next examined whether increased BDNF production would lead to activation of TrkB, which was assessed by its tyrosine phosphorylation [61]. A single leptin injection (5 mg/kg, i.p.) increased phosphorylated TrkB levels in the hippocampus at 2 and 6 h post injection (2 h, t(9) = 2.3740, P = 0.0417; 6 h, t(8) = 2.4050, P = 0.0428) (Fig. 2c), suggesting that leptin induces activation of the BDNF-TrkB signaling pathway.
Regulation of exon-specific Bdnf mRNA expression by leptin treatment, leptin deficiency and leptin receptor deficiency
Leptin-induced changes in total (exon IX) Bdnf mRNA levels could be related to the changes in noncoding exons I–VIII spliced to exon IX (Fig. 3a). To determine which noncoding exons may directly contribute to the modulation of total Bdnf expression by leptin, we analyzed exons I, II, III, IV, and VI that are highly expressed in the hippocampus. Exons V, VII, and VIII were not included due to their very low expression levels (data not shown). While mRNA levels of Bdnf exons II and III remained unchanged, exons I, IV, and VI transcripts were found to increase 2 h after leptin treatment (5 mg/kg) (treatment: F(1,155) = 52.8300, P < 0.001; exon: F(4,155) = 2.8020, P = 0.0278; treatment × exon interaction: F(4,155) = 2.8020, P = 0.0278; exons I: P < 0.001; exons II: P = 0.0511; exons III: P > 0.9999; exons IV: P < 0.001; exons VI: P < 0.001) (Fig. 3b). These results suggest that Bdnf transcripts containing exons I, IV, and VI are responsible for leptin-induced upregulation of total Bdnf gene expression in the hippocampus.
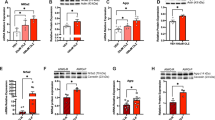
a The structure of the mouse Bdnf gene. b Wild-type mice. Exon-specific Bdnf mRNA expression levels. Saline, n = 16; leptin (5 mg/kg, i.p.), n = 17. c ob/ob mice. Upper panel: left, schematic diagram of breeding strategy; middle, total Bdnf mRNA; right, Bdnf exon-specific mRNA. WT, n = 6, ob/ob, n = 6. Lower panel: left, schematic diagram of breeding strategy and leptin treatment; middle, total Bdnf mRNA; right, Bdnf exon-specific mRNA. n = 5 per group. d db/db mice. Left, schematic diagram of breeding strategy; middle, total Bdnf mRNA; right, Bdnf exon-specific mRNA. WT, n = 5; db/db, n = 5. e Lepr conditional knockout mice. Left, schematic diagram of breeding strategy; middle, total Bdnf mRNA; right, Bdnf exon-specific mRNA. Leprflox/flox, n = 7; Leprflox/flox;Emx1-Cre, n = 7. *P < 0.05; **P < 0.01; ***P < 0.001 compared with wild-type littermates or saline-injected controls.
We then asked whether deficiency of leptin or the leptin receptor represses Bdnf mRNA expression. We investigated the modulation of total Bdnf mRNA (exon IX) in the hippocampus as compared to mRNA levels of noncoding exons. Leptin-deficient (ob/ob) mice showed decreased total Bdnf mRNA expression (t(10) = 3.0790, P = 0.0117) (Fig. 3c). Analysis of exon-specific Bdnf expression revealed that mRNAs for exons II, III, IV, and VI were downregulated in ob/ob mice (genotype: F(1,50) = 31.0300, P < 0.001; exon: F(4,50) = 0.7510, P = 0.5620; genotype × exon interaction: F(4,50) = 0.7510, P = 0.5620, exons I: P > 0.9999; exons II: P = 0.0305; exons III: P = 0.0412; exons IV: P = 0.0303; exons VI: P = 0.0197) (Fig. 3c). We next determined whether leptin replenishment in ob/ob mice can restore abnormal Bdnf expression. Ob/ob mice and WT littermates received a single i.p. injection of leptin (5 mg/kg) 2 h prior to dissection of hippocampal tissue. Leptin replacement rescued the downregulated expression of total Bdnf mRNA in ob/ob mice (P = 0.0381), comparable to the level seen in WT littermates (P = 0.0205) (genotype: F(1,16) = 27.0900, P < 0.001; treatment: F(1,16) = 21.5900, P < 0.001; genotype × treatment interaction: F(1,16) = 0.0433, P = 0.8377) (Fig. 3c). mRNA expression of exons IV and VI, but not exons I, II, and III, was restored by leptin (treatment: F(1, 80) = 29.7200, P < 0.001; genotype: F(1,80) = 57.6200, P < 0.001; exon: F(4,80) = 6.7280, P < 0.001; genotype × treatment × exon interaction: F(4,80) = 2.3570, P = 0.0605) (Fig. 3c). The failure of leptin to reverse the downregulation of exons I, II, and III suggests that their repression is not a direct result of leptin deficiency.
The above results indicate that leptin deficiency decreases total Bdnf mRNA expression via downregulation of specific Bdnf exons. To confirm that lack of LepRb would exert similar effects, we analyzed Bdnf gene expression in db/db mice that lack intracellular leptin action through LepRb [55, 62]. As seen in ob/ob mice, total Bdnf mRNA (exon IX) expression was decreased in db/db mice (t-test with Welch’s correction, P = 0.0477) (Fig. 3d). Among other exons detected, mRNAs for exons I, III, and IV were downregulated (Fig. 3d) (genotype: F(1,40) = 35.7800, P < 0.001; exon: F(4,40) = 0.9118, P = 0.4664; treatment × exon interaction: F(4,40) = 1.0480, P = 0.3949, exons I: P = 0.0015; exon II: P = 0.2189; exon III: P = 0.0183; exon IV: P = 0.0268; exon VI: P > 0.9999), which is distinct from that observed in ob/ob mice.
A global loss of leptin signaling in ob/ob and db/db mice causes obesity and type 2 diabetes [1, 8], which could contribute to the downregulation of Bdnf mRNA expression in the hippocampus. We have previously shown that forebrain-specific LepRb knockout (Leprflox/flox;Emx1-Cre) mice have normal body weight and insulin levels [13], which have decreased LepRb mRNA expression in the hippocampus compared to WT (Leprflox/flox: 1.00 ± 0.0778; Leprflox/flox;Emx1-Cre: 0.46 ± 0.0695; t(12) = 5.2030, P < 0.001). Therefore, we used Leprflox/flox;Emx1-Cre mice to examine the effect of loss of LepRb on Bdnf gene expression. In contrast to ob/ob and db/db mice showing increased body weight, we confirmed that Leprflox/flox;Emx1-Cre mice had normal body weight (Leprflox/flox: 26.86 ± 0.349 g; Leprflox/flox;Emx1-Cre: 26.93 ± 0.308 g; t(12) = 0.1534, P = 0.8807). Total Bdnf mRNA expression was reduced in Leprflox/flox;Emx1-Cre mice (t(12) = 3.7760, P = 0.0026) (Fig. 3e), accompanied by distinct exon-specific expression patterns with downregulation of Bdnf exon I, II, IV, and VI expression (genotype: F(1,60) = 44.6700, P < 0.001; exon: F(4,60) = 1.2560, P = 0.2971; treatment × exon interaction: F(4,60) = 1.2560, P = 0.2971, exon I: P < 0.001; exon II: P = 0.0192; exon III: P > 0.9999; exon IV: P = 0.0012; exon VI: P = 0.0391) (Fig. 3e). These results indicate that the three lines of mutant mice lacking leptin signaling displayed a similar reduction of total Bdnf mRNA expression but different exon-specific downregulation patterns.
All experiments described above were conducted in male mice. We also examined the effects of leptin and deficiency in leptin or LepRb on Bdnf gene expression in female mice. As we have previously demonstrated that leptin induces AKT phosphorylation and antidepressant-like effects in the proestrus phase but not in the diestrus phase [14], we therefore tested the effects of leptin on Bdnf gene expression during these two stages of the estrous cycle. The estrous cycle was identified based on vaginal lavage prior to the treatments. Female mice in proestrus or diestrus received an i.p. injection with either vehicle (saline) or leptin (5.0 mg/kg), and 2 h later hippocampal tissue was collected for the measurement of mRNA levels. Leptin increased total Bdnf mRNA expression in the proestrus phase but not in the diestrus phase (treatment: F(1,40) = 8.3640, P = 0.0062; estrus cycle: F(1,40) = 0.9669, P = 0.3314; treatment × estrus cycle interaction: F(1,40) = 10.3400, P = 0.0026) (Fig. 4a). Correspondingly, mRNA levels for exons I, II, III, and VI were increased in proestrus female mice but not in diestrus female mice (exon I: treatment: F(1,40) = 12.4800, P = 0.0011; estrus cycle: F(1,40) = 1.0630, P = 0.3087; treatment × estrus cycle interaction: F(1,40) = 4.5870, P = 0.0384; exon II: treatment: F(1,40) = 11.7200, P = 0.0014; estrus cycle: F(1,40) = 0.1099, P = 0.7420; treatment × estrus cycle interaction: F(1,40) = 6.6790, P = 0.0135; exon III: treatment: F(1,40) = 4.0010, P = 0.0523; estrus cycle: F(1,40) = 0.0719, P = 0.7899; treatment × estrus cycle interaction: F(1,40) = 7.9640, P = 0.0074; exon IV: treatment: F(1,40) = 0.0296, P = 0.8642; estrus cycle: F(1,40) = 3.4420, P = 0.0709; treatment × estrus cycle interaction: F(1,40) = 3.9090, P = 0.0550; exon IV: treatment: F(1,40) = 0.0296, P = 0.8642; estrus cycle: F(1,40) = 3.4420, P = 0.0709; treatment × estrus cycle interaction: F(1,40) = 3.9090, P = 0.0550; exon VI: treatment: F(1,40) = 7.0160, P = 0.0115; estrus cycle: F(1,40) = 4.7560, P = 0.0351; treatment × estrus cycle interaction: F(1,40) = 2.7830, P = 0.1031) (Fig. 4a). Furthermore, mRNA levels for total Bdnf and exons IV and VI varied across the estrous cycle with higher levels in the proestrus phase (total Bdnf, P = 0.0301; exons I: P = 0.8960; exons II: P = 0.9686; exons III: P = 0.2856; exon IV, P = 0.0467; exon VI, P = 0.0454) (Fig. 4a). These results suggest that basal expression levels of total Bdnf and specific exons and their responses to leptin treatment are estrous cycle-dependent, which could be partially due to a dynamic change in endogenous leptin and LepRb across the estrous cycle [63].
a Wild-type female mice. Upper panel, schematic diagram depicting the identification of the estrous cycle and leptin treatment. Middle and lower panels, total Bdnf and exon-specific mRNA expression levels in proestrus versus diestrus in response to leptin injection (5 mg/kg, i.p.). n = 11 per group. b ob/ob female mice. Left, schematic diagram of breeding strategy; middle, total Bdnf mRNA; right, Bdnf exon-specific mRNA. n = 6 per group. c db/db female mice. Left, schematic diagram of breeding strategy; middle, total Bdnf mRNA; right, Bdnf exon-specific mRNA. n = 6 per group. *P < 0.05; **P < 0.01; ***P < 0.001 compared with WT or saline-injected controls.
Both ob/ob and db/db female mice have impaired ovarian development [64]. They are infertile and exhibit no evidence of estrous cycles [65]. As seen in male mice, total Bdnf mRNA expression in the hippocampus was decreased in female ob/ob and db/db mice (ob/ob: t(10) = 2.6750, P = 0.0233; db/db: t(10) = 2.2510, P = 0.0481) with overlapping but distinct profiles of specific exon mRNA expression (Fig. 4b, c). While mRNAs for exons I, III, and VI were decreased in ob/ob female mice (genotype: F(1,50) = 33.3600, P < 0.001; exon: F(4,50) = 1.0160, P = 0.4083; genotype × exon interaction: F(4,50) = 1.0160, P = 0.4083) (Fig. 4b), db/db female mice showed decreased levels of exons I, III, IV, and VI mRNAs (Fig. 4b, c) (genotype: F(1,50) = 15.1900, P < 0.001; exon: F(4,50) = 2.5860, P = 0.0480; genotype × exon interaction: F(4,50) = 2.5860, P = 0.0480). These data suggest that the effects of leptin deficiency and leptin receptor deficiency on the transcription of Bdnf gene expression are sex-dependent.
Leptin regulates Bdnf gene expression through epigenetic mechanisms
We hypothesized that epigenetic modifications of the promoter region of Bdnf exons underlie leptin-induced regulation of Bdnf gene expression. To test this hypothesis, we first examined whether histone modifications at specific Bdnf promoters are altered in the hippocampus following leptin treatment. ChIP assays were performed at 2 h after leptin injection (5 mg/kg, i.p.) to quantify the relative levels of acetylated H3 and H4 at the promoter of exons I, II, III, IV, and VI (Fig. 5a). Histone H3 acetylation was greatly increased at promoters I, IV, and VI (treatment: F(1,48) = 21.8300, P < 0.001; promoter: F(5,48) = 1.5430, P = 0.3558; treatment × promoter interaction: F(5,48) = 1.5430, P = 0.1942, promoter I: P = 0.0325; promoter II: P = 0.4199; promoter III: P > 0.9999; promoter IV: P = 0.0256; promoter VI: P = 0.0377; GAPDH: P > 0.9999), while no change in histone H4 acetylation was detected at any promoter (treatment: F(1,48) = 0.7230, P = 0.3994; promoter: F(5,48) = 0.1289, P = 0.9851; treatment × promoter interaction: F(5,48) = 0.1289, P = 0.9851, promoter I: P > 0.9999; promoter II: P > 0.9999; promoter III: P > 0.9999; promoter IV: P > 0.9999; promoter VI: P > 0.9999; GAPDH: P > 0.9999) (Fig. 5a). In addition, we examined methylation of lysine 4, 9, and 27 on histone H3 at the Bdnf promoters. Leptin increased dimethylation of H3K4 at promoters I, IV, and VI (treatment: F(1,84) = 41.0100, P < 0.001; promoter: F(5,84) = 2.5460, P = 0.0340; treatment × promoter interaction: F(5,84) = 2.5460, P = 0.0340, promoter I: P = 0.0062; promoter II: P > 0.9999; promoter III: P = 0.6081; promoter IV: P < 0.001; promoter VI: P = 0.0063; GAPDH: P = 0.9830), decreased dimethylation of H3K9 at promoters II, IV, and VI (treatment: F(1,84) = 17.9800 P < 0.001; promoter: F(5,84) = 1.6440, P = 0.1573; treatment × promoter interaction: F(5,84) = 1.7060, P = 0.1422, promoter I: P > 0.9999; promoter II: P = 0.0348; promoter III: P > 0.9999; promoter IV: P = 0.0406; promoter VI: P = 0.0199; GAPDH: P > 0.9999), and had no effect on demethylation of H3K27 at any promoter (treatment: F(1,84) = 0.3343, P = 0.5647; promoter: F(5,84) = 0.3451, P = 0.8841; treatment × promoter interaction: F(5,84) = 0.3451, P = 0.8841, promoter I: P > 0.9999; promoter II: P > 0.9999; promoter III: P > 0.9999; promoter IV: P > 0.9999; promoter VI: P > 0.9999; GAPDH: P > 0.9999) (Fig. 5a).
a Upper left, schematic diagram of histone modifications. Upper middle, Histone H3 acetylation (n = 5 per group); upper right, histone H4 acetylation (n = 5 per group); lower left, H3K4 methylation (n = 8 per group); lower middle, H3K9 methylation (n = 8 per group); H3K27 methylation (n = 8 per group). b Upper panel, schematic diagram of the positions of Bdnf CpG islands 1–6. Middle and lower panels, percentage of methylation of each CpG site. DNA methylation levels of CpG1–CpG6, n = 5 per group. *P < 0.05; **P < 0.01; ***P < 0.001 compared with saline-injected controls.
Alternatively, DNA methylation changes in the Bdnf gene region might mediate leptin-induced regulation of Bdnf gene expression in the hippocampus [66, 67]. Thus, we chose the CpG island of the Bdnf gene upstream of exon I (CpG1) and CpG islands within the promoters II, IV, and VI and the exonic regions of the Bdnf gene after the transcriptional start site of exons II, IV, and VI (CpG2–6) as targets for methylation analysis. Using bisulfite pyrosequencing, the methylation levels of CpG sites at Bdnf promoters/exon regions were quantified. No significant changes in methylated DNA levels associated with exons I, IV, and IV were observed after leptin treatment compared with vehicle controls (Fig. 5b), except for one out of 13 CpG sites within the promoter II CpG island, i.e., CpG island 2 (CpG1: treatment: F(1,88) = 1.5180, P = 0.2211; CpGs: F(10,88) = 37.9300, P < 0.001; treatment × CpG interaction: F(10,88) = 0.5878, P = 0.8199; CpG2: treatment: F(1,104) = 4.6230, P = 0.0339; CpGs: F(12,104) = 7.4440, P < 0.001; treatment × CpG interaction: F(12,104) = 2.3690, P = 0.0097; CpG3: treatment: F(1,96) = 0.4993, P = 0.4815; CpGs: F(11,96) = 18.1600, P < 0.001; treatment × CpG interaction: F(11,96) = 0.7055, P = 0.7305; CpG4: treatment: F(1,64) = 0.0011, P = 0.9740; CpGs: F(7,64) = 83.0200, P < 0.001; treatment × CpG interaction: F(7,64) = 0.7014, P = 0.6707; CpG5: treatment: F(1,136) = 0.0775, P = 0.7812; CpGs: F(16,136) = 3.0240, P < 0.001; treatment × CpG interaction: F(16,136) = 1.0210, P = 0.4392; CpG6: treatment: F(1,120) = 0.0880, P = 0.7673; CpGs: F(14,120) = 12.3800, P < 0.001; treatment × CpG interaction: F(14,120) = 0.7632, P = 0.7065). Our data indicate that the methylation of 75 out of 76 CpG sites that we analyzed was not sensitive to leptin treatment.
Leptin-induced regulation of Bdnf mRNA expression is mediated by stimulation of an AKT/p300 HAT/H3 modification cascade
The binding of leptin to LepRb activates three major signaling pathways, i.e., JAK2/STAT3, PI3K/AKT, and ERK pathways [68] (Fig. 6a). To explore which signaling pathway(s) is responsible for mediating leptin’s effects on Bdnf gene expression, we assessed activation of STAT3, AKT, and ERK in the hippocampus by leptin. Leptin treatment at a dose used for gene expression (i.e., 5 mg/kg, i.p.) failed to induce a change in phosphorylation levels of STAT3 or ERK (t(7) = 0.5803, P = 0.5782), but significantly increased AKT phosphorylation (t(7) = 3.8020, P = 0.0067), suggesting that leptin action in the hippocampus may be mediated by AKT signaling (Fig. 6a). Indeed, we have previously shown that AKT activation in the hippocampus mediates leptin’s antidepressant-like behavioral effects [14]. To examine whether AKT signaling is required for leptin-induced Bdnf gene expression, an AKT selective inhibitor, AKTi, was infused into the dentate gyrus to block AKT signaling prior to leptin injection. We found that inhibition of AKT activation eliminated the effects on total Bdnf mRNA expression induced by leptin (Kruskal–Wallis test, P < 0.001) (Fig. 6b). In addition, pretreatment with AKTi blocked the effects of leptin on acetylation of H3 at promoters I, IV, and VI and methylation of H3K4 at promoters I, IV, and VI and H3K9 at promoters II, IV, and VI (Fig. 6c) (H3, treatment: F(2,138) = 12.1000, P < 0.001; promoter: F(5,138) = 3.7980, P = 0.0030; treatment × promoter interaction: F(10,138) = 2.1530, P = 0.0242); H3K4, treatment: F(2,138) = 20.4300, P < 0.001; promoter: F(5,138) = 1.2300, P = 0.2982; treatment × promoter interaction: F(10,138) = 1.9550, P = 0.0428; H3K9, treatment: F(2,138) = 11.9700, P < 0.001; promoter: F(5,138) = 1.5530, P = 0.1774; treatment × promoter interaction: F(10,138) = 1.5140, P = 0.1403. These results suggest that the PI3K/AKT pathway plays a critical role in leptin-induced regulation of Bdnf gene expression.
a Left, schematic representation of leptin-mediated signaling pathways; middle: western blotting images showing phosphorylation of AKT, ERK, and STAT3 in the hippocampus induced by leptin; right, quantitative data from western blots. Saline, n = 5; leptin, n = 4. Blockade of leptin-induced increases in total Bdnf mRNA expression (b) and changes in H3 acetylation, H3K4 methylation, and H3K9 methylation (c) and p300 HAT binding by the AKT inhibitor AKTi (d). Vehicle-saline, n = 9; vehicle + leptin, n = 9; AKTi + leptin, n = 8. Blockade of leptin-induced increases in total Bdnf mRNA expression (e), H3 acetylation (f), H3K4 and H3K9 methylation (g) by the p300 HAT inhibitor C646. n = 6 per group. *P < 0.05; **P < 0.01; ***P < 0.001 compared with saline-treated controls.
AKT can phosphorylate p300 HAT at Ser-1834, thereby increasing its promoter recruitment and histone acetylation [69]. We then examined whether AKT activation is involved in p300 HAT binding activity at Bdnf promoters. Leptin was found to increase p300 HAT binding to Bdnf promoters I, IV, and VI, and this effect was abolished by pretreatment with AKTi in the hippocampus (treatment: F(2,138) = 14.2600, P < 0.001; promoter: F(5,138) = 4.0820, P = 0.0017; treatment × promoter interaction: F(10,138) = 2.5300, P = 0.0079) (Fig. 6d). Next, we tested whether p300 HAT activity is necessary for leptin-induced changes in Bdnf gene expression. C646, a competitive p300 HAT inhibitor [70], was injected into the dentate gyrus prior to leptin treatment. Pretreatment with C646 was sufficient to block leptin-induced increase in total Bdnf mRNA expression (F(2,15) = 6.9240, P = 0.0074) (Fig. 6e) and subsequent acetylation of histone H3 at promoters I, IV, and VI was abolished (treatment: F(2,90) = 9.8950, P < 0.001; promoter: F(5,90) = 2.2800, P = 0.0532; treatment × promoter interaction: F(10,90) = 2.1570, P = 0.0277) (Fig. 6f), suggesting that p300 HAT mediates histone acetylation at Bdnf promoters. p300 HAT-mediated histone acetylation events have been shown to modulate H3 methylation [71]. We therefore examined whether methylation of H3K4 and H3K9 at Bdnf promoters is affected by p300 HAT activity. Pretreatment with C646 was found to abolish leptin-induced increase in H3K4 methylation and decrease in H3K9 methylation (H3K4, treatment: F(2,90) = 12.1900, P < 0.001; promoter: F(5,90) = 0.9871, P = 0.4302; treatment × promoter interaction: F(10,90) = 1.1900, P = 0.3083; H3K9, treatment: F(2,90) = 10.7300, P < 0.001; promoter: F(5,90) = 0.8255, P = 0.5348; treatment × promoter interaction: F(10,90) = 1.1440, P = 0.3393) (Fig. 6g), suggesting p300 HAT activity is also indirectly involved in histone H3 lysine methylation. Together, these results suggest that leptin induces Bdnf gene expression through an AKT/p300 HAT/H3 modification cascade, leading to chromatin remodeling and transcriptional activation.
Bdnf gene expression in the dentate gyrus is essential for the antidepressant-like effect of leptin
We next asked whether Bdnf gene expression in the dentate gyrus is required for leptin’s depression-related behaviors. To address this question, AAV-Cre and AAV-GFP control vectors were injected into the dentate gyrus of the hippocampus of Bdnfflox/flox mice (Fig. 7a). Three weeks later, the expression of Cre expression and the deletion of Bdnf exon IX in the dentate gyrus were confirmed (Bdnf exon IX: Mann–Whitney test, P < 0.001) (Fig. 7a). AAV-Cre and AAV-GFP-treated mice received i.p. injection of leptin (1 or 5 mg/kg, i.p.) 2 h before the forced swim and tail suspension tests. Leptin decreased immobility time in both tests in AAV-GFP treated control mice; however, this effect was eliminated in AAV-Cre treated mice (Fig. 7a) (forced swim test, 1 mg/kg, 2 h: treatment: F(1,33) = 12.2700, P = 0.0013; genotype: F(1,33) = 8.7090, P = 0.0058; genotype × treatment: F(1,33) = 4.1910, P = 0.0487; tail suspension test, 5 mg/kg, 2 h: treatment: F(1,33) = 5.9060, P = 0.0207; genotype: F(1,33) = 5.3540, P = 0.0270; genotype × treatment: F(1,33) = 7.5930, P = 0.0095). This insensitivity to leptin is unlikely to be due to a downregulation of LepRb in the dentate gyrus caused by deletion of Bdnf, as LepRb mRNA levels were unchanged in AAV-Cre treated mice (Mann–Whitney test, P = 0.5567) (Fig. 7a). Moreover, the deletion of Bdnf in the dentate gyrus had no effect on body weight (t(35) = 0.8378, P = 0.4078) (Fig. 7a).
a Dentate gyrus (DG)-specific deletion of Bdnf. Upper left, schematic diagram of the Bdnf floxed allele after AAV-Cre-mediated recombination. Upper middle-left, stereotaxic injection of AAV-Cre-GFP or AAV-GFP vectors in the DG of Bdnfflox/flox mice; upper middle-right, Cre mRNA expression; upper right, total Bdnf mRNA expression in the DG. Lower left, forced swim test (FST); lower middle-left, tail suspension test (TST); lower middle-right, LepRb mRNA expression in the DG; lower right, body weight. AAV-GFP + saline, n = 10; AAV-GFP + leptin, n = 10; AAV-Cre + saline, n = 9; AAV-GFP + leptin, n = 8. Total AAV-GFP, n = 20; total AAV-Cre, n = 17. b Left panel, schematic diagram of breeding strategy. Upper panel, Cre (left), total Bdnf (middle), and LepRb (right) mRNA expression in the DG. n = 5 per group. Lower left, forced swim test; Bdnfflox/flox + saline, n = 11, Bdnfflox/flox + leptin, n = 11, Bdnfflox/flox;Lepr-ires-Cre + saline, n = 11, Bdnfflox/flox;Lepr-ires-Cre + leptin, n = 12. Lower middle, body weight; Bdnfflox/flox, n = 22, Bdnfflox/flox;Lepr-ires-Cre, n = 23. Lower right, total Bdnf mRNA level 2 h after leptin injection (5 mg/kg, i.p.). n = 5 per group. *P < 0.05; ** P < 0.01; *** P < 0.001 compared with WT littermates or saline-treated controls.
Given that LepRb neurons expressed Bdnf in the dentate gyrus, we asked whether an intracellular interaction between LepRb and Bdnf mediates leptin action on despair behavior. To address this, Lepr-ires-Cre mice were crossed with Bdnfflox/flox mice to induce deletion of Bdnf specifically in LepRb neurons (Fig. 7b). The resultant Bdnfflox/flox;Lepr-ires-Cre mice confirmed Cre expression in the dentate gyrus with a dramatic reduction of Bdnf expression (t(8) = 2.7830, P = 0.0238) but normal levels of LepRb mRNA (t(8) = 0.0925, P = 0.9286) (Fig. 7b). Bdnf flox/flox;Lepr-ires-Cre mice and Bdnfflox/flox littermate controls were injected with leptin (5 mg/kg, i.p.) 2 h before the forced swim test. We found that the targeted deletion of Bdnf in LepRb neurons abolished leptin’s antidepressant-like effects in Bdnfflox/flox;Lepr-ires-Cre mice (treatment: F(1,41) = 23.5300, P < 0.001; genotype: F(1,41) = 0.8602, P = 0.3591; genotype × treatment: F(1,41) = 5.7670, P = 0.0210) (Fig. 7b). As seen in mice with the deletion of Bdnf in the dentate gyrus, body weight was unaltered in Bdnf flox/flox,Lepr-ires-cre mice (t(43) = 0.4250, P = 0.6729) (Fig. 7b). Lastly, we tested whether leptin-induced Bdnf gene expression is dependent on Bdnf expression in LepRb neurons. Total Bdnf mRNA levels were measured 2 h after leptin treatment (5 mg/kg, i.p.). We showed that leptin-induced increase in total Bdnf mRNA expression was eliminated by deletion of Bdnf in LepRb neurons (treatment: F(1,16) = 8.0840, P = 0.0117; genotype: F(1,16) = 5.8620, P = 0.0277; genotype × treatment: F(1,16) = 3.9460, P = 0.0644) (Fig. 7b), suggesting that leptin regulates Bdnf gene expression via a direct mechanism.
Discussion
The hippocampus is a highly vulnerable, dynamic, and responsive brain structure in that it demonstrates rapid plasticity at the molecular, cellular, circuit, and functional levels [72, 73]. BDNF, which is expressed at the highest levels in the hippocampus, plays a vital role in hippocampal plasticity and function [74, 75]. In this study, our finding that dentate gyrus neurons expressing the functional leptin receptor isoform LepRb also contained BDNF suggests that BDNF-expressing neurons are targets of leptin action. Further studies showed that leptin upregulated total Bdnf gene expression and protein synthesis, causing activation of TrkB in the hippocampus. Bdnf transcription is uniquely controlled by nine individual promoters, which drive expression of multiple transcripts encoding the same protein [34,35,36]. Such complex gene structure suggests that Bdnf gene expression is finely tuned. We found that leptin treatment increased transcriptional activity of Bdnf promoters I, IV, and VI in the hippocampus. However, leptin deficiency or loss of its receptor decreased Bdnf gene expression, albeit via suppression of transcriptional activity at different promoters. Furthermore, leptin activated AKT signaling, which in turn regulated p300 HAT recruitment and subsequently promoted the transcriptional activity of Bdnf promoters via an epigenetic mechanism. Importantly, the loss of Bdnf gene expression in the dentate gyrus eliminated the antidepressant-like behavioral effects of leptin, suggesting that upregulation of BDNF underlies leptin’s action on mood-related behavior.
BDNF in the hippocampus has been implicated in the pathophysiology of depression and the therapeutic mechanisms of antidepressant treatments [23, 47, 76]. In animal studies, BDNF levels in the hippocampus decrease in response to a variety of stressors and increase following various antidepressant treatments [76]. Postmortem studies demonstrate that levels of BDNF in the hippocampus (both mRNA and protein) are reduced in depressed patients [77, 78], whereas BDNF levels are elevated in patients receiving antidepressants [26, 77,78,79,80]. Moreover, overexpressing BDNF in excitatory neurons of the forebrain (including hippocampus) caused an antidepressant-like behavioral phenotype [81], while a single infusion of BDNF directly into the hippocampus was sufficient to induce an antidepressant-like effect [29]. Importantly, loss of forebrain BDNF or reduced BDNF signaling (overexpression of truncated TrkB) attenuates the antidepressant-like response to antidepressants [27, 82,83,84]. The rapid acting antidepressant ketamine is also dependent on increased BDNF signaling [30, 85]. Furthermore, mice with reduced BDNF signaling caused by overexpressing truncated TrkB are resistant to the effects of antidepressants [27]. We have shown that leptin has antidepressant-like effects mediated by LepRb in the hippocampus [13,14,15, 22, 53]. Our findings that leptin increased Bdnf gene expression, while the loss of BDNF abolished leptin’s antidepressant-like effects, support the idea that hippocampal BDNF is required for leptin action on mood-related behavior. Furthermore, this effect is likely to be mediated by direct interactions between LepRb and BDNF, as deletion of Bdnf from LepRb neurons in the dentate gyrus eliminated the antidepressant-like effects of leptin.
Bdnf gene expression is regulated by diverse stimuli and physiological and pathological conditions through differential recruitment of individual exon-specific Bdnf promoters. Bdnf transcripts containing exons I, II, III, IV, and VI are present in the hippocampus, and they make differential contributions to total BDNF [86]. Many antidepressants have been shown to result in distinct exon-specific Bdnf transcripts [44, 87,88,89]. For example, tranylcypromine and desipramine increase exon I, II, and IV (original III) mRNAs, fluoxetine treatment enhances activity of promoter II [88], whereas reboxetine increases activation of promoter IV [87]. Leptin, similar to classic antidepressants, increased total Bdnf gene expression while inducing sexually dimorphic profiles of Bdnf promoter activation, with male mice exhibiting increased expression of exons I, IV, and VI and female mice exhibiting increased expression of exons I, II, III, and VI. In contrast, leptin deficiency or leptin receptor deficiency decreased total Bdnf gene expression. However, three distinct lines of male mice deficient in leptin or leptin signaling, i.e., ob/ob, db/db and Leprflox/flox;Emx1-Cre mice, exhibited different patterns of downregulation of exon-specific Bdnf gene expression, with ob/ob associated with decreased expression of exons II, III, IV, and VI, db/db associated with decreased expression of exons I, III, and IV and Leprflox/flox;Emx1-Cre associated with decreased expression of exons I, II, IV, and VI. In ob/ob mice, replenishing leptin restored the levels of transcriptional activity at promoters IV and VI but failed to reverse the downregulation of exons II and III. These results suggest that leptin deficiency is responsible for the downregulation of exons IV and VI expression, but not exons II and III. We considered the possibility that the downregulated expression of exons II and III in ob/ob mice was caused by obesity, which cannot be normalized by acute leptin treatment. This seems to be the case for exon III but not for exon II, as expression of this exon was also decreased in Leprflox/flox;Emx1-Cre mice with normal body weight. Bdnf regulation appears to be even more complicated in female mice. First, leptin increases the transcription of the Bdnf gene in proestrus but not diestrus, with specific changes in expression of exons I, II, III, and VI. Second, while leptin deficiency caused a reduction of expression of exons I, III, and VI in female ob/ob mice, female db/db mice exhibited decreased expression levels of exons I, III, IV, and VI. Bdnf promoters show differential transcription efficiency with promoters IV and VI contributing more substantially to total BDNF in the hippocampus [86]. How the different patterns of Bdnf exon-specific upregulation or downregulation induced by leptin, leptin deficiency and leptin receptor deficiency contribute to total BDNF are dependent on the relative abundance of individual Bdnf transcripts and the fold changes of their relative abundance following different manipulations. Our results demonstrate that transcription of the Bdnf gene is tightly regulated by leptin, displaying distinct exon-specific, sex-dependent expression profiles in response to elevated leptin levels, defective leptin signaling, and leptin resistance. These findings suggest that Bdnf expression in the hippocampus is sensitive to fluctuations in leptin availability and leptin signaling, serving as a finely tuned neural component of the adipose-brain axis.
One intriguing and important finding of this study is that leptin activates the PI3K/AKT pathway to recruit the p300 HAT, leading to histone modifications and subsequently transcriptional activation of the Bdnf gene (Fig. 8). This finding constitutes strong evidence for an intersection between cell signaling and epigenetic regulation. First, among three major signaling pathways downstream of LepRb, the AKT pathway was selectively activated in the hippocampus by leptin. Moreover, we have previously shown that AKT signaling in the hippocampus is required for leptin’s antidepressant-like behavioral effects [14]. Second, AKT can phosphorylate p300 HAT at Ser-1834 [69], which could induce its recruitment to the Bdnf promoter, leading to the acetylation of histones and activation of the transcriptional machinery. Indeed, leptin treatment increased p300 HAT binding to Bdnf promoters I, IV, and VI, and this effect was attenuated by inhibition of AKT. Furthermore, C646, a p300 HAT inhibitor, attenuated leptin-induced acetylation of histone H3. AKT can also phosphorylate other chromatin-modifying enzymes, such as the histone methyltransferase EZH2 (H3K27 methylation) and DNA methyltransferase DNMT1, resulting in changes in function of these enzymes [90, 91]. Unlike histone acetylation, which occurs rapidly in response to stimuli, histone methylation, and DNA methylation are relatively static [90]. In this study, we analyzed the methylation level of 76 CpG sites (6 CpG islands) within the promoters I, II, IV, and VI and the exonic regions following leptin treatment. The methylation at 75 CpG sites was unaltered by acute leptin treatment, suggesting that DNA methylation may not play a significant role in leptin-induced Bdnf gene expression. However, we cannot rule out the possibility that other CpG sites we did not analyze may be sensitive to leptin treatment. Methylation of histones at different residues results in different transcriptional outcomes. For example, methylation of H3K27 and H3K9 is associated with transcriptional repression, whereas methylation on H3K4 usually increases gene expression [92,93,94]. We observed increased H3K4 methylation coupled with decreased H3K9 methylation with no changes in H3K27 methylation at the Bdnf promoter following leptin treatment. Interestingly, blockade of p300 HAT with C646 also abolished leptin-induced changes in methylation of H3K4 and H3K9. The involvement of p300 HAT in the regulation of histone H3 methylation at the Bdnf promoter may be secondary to its effect on histone H3 acetylation. In support of this, one study reported that H3K4 methylation is dependent upon p300 HAT-mediated H3 acetylation [71]. The methyltransferase SET1C, which methylates histone H3 at lysine 4, was demonstrated to act synergistically with p300 HAT through direct interactions and coupled histone modifications [71]. Our results support that direct communications between the AKT signaling pathway and the chromatin-modifying machinery mediates Bdnf gene expression in response to leptin. This may represent a generalized epigenetic mechanism that permits cells to respond dynamically to environmental signals.
Our findings support the idea that leptin interacts with BDNF via LepRb in the hippocampus to exert its behavioral effects, beyond its role as an adiposity signal. Although the present study only investigated leptin’s antidepressant-like effects, the interactions between leptin signaling and BDNF hypothetically could extend to other hippocampal functions, such as learning and memory [18, 95, 96]. Moreover, regulation of Bdnf gene expression by leptin signaling is complex, with differential regulation of exon-specific expression in a sex-dependent manner. BDNF, in turn, is required for leptin action in the hippocampus. Thus, dysregulation of BDNF presumably could lead to resistance to leptin, contributing to the pathogenesis of depression and Alzheimer’s disease.
References
Zhang Y, Proenca R, Maffei M, Barone M, Leopold L, Friedman JM. Positional cloning of the mouse obese gene and its human homologue. Nature. 1994;372:425–32.
Halaas JL, Gajiwala KS, Maffei M, Cohen SL, Chait BT, Rabinowitz D, et al. Weight-reducing effects of the plasma protein encoded by the obese gene. Science. 1995;269:543–6.
Golden PL, Maccagnan TJ, Pardridge WM. Human blood-brain barrier leptin receptor. Binding and endocytosis in isolated human brain microvessels. J Clin Investig. 1997;99:14–8.
Scott MM, Lachey JL, Sternson SM, Lee CE, Elias CF, Friedman JM, et al. Leptin targets in the mouse brain. J Comp Neurol. 2009;514:518–32.
Patterson CM, Leshan RL, Jones JC, Myers MG Jr. Molecular mapping of mouse brain regions innervated by leptin receptor-expressing cells. Brain Res. 2011;1378:18–28.
Elias CF, Kelly JF, Lee CE, Ahima RS, Drucker DJ, Saper CB, et al. Chemical characterization of leptin-activated neurons in the rat brain. J Comp Neurol. 2000;423:261–81.
Baumann H, Morella KK, White DW, Dembski M, Bailon PS, Kim H, et al. The full-length leptin receptor has signaling capabilities of interleukin 6-type cytokine receptors. Proc Natl Acad Sci USA. 1996;93:8374–8.
Tartaglia LA, Dembski M, Weng X, Deng N, Culpepper J, Devos R, et al. Identification and expression cloning of a leptin receptor, OB-R. Cell. 1995;83:1263–71.
Bjorbaek C, Elmquist JK, Michl P, Ahima RS, van Bueren A, McCall AL, et al. Expression of leptin receptor isoforms in rat brain microvessels. Endocrinology. 1998;139:3485–91.
Friedman J. The long road to leptin. J Clin Investig. 2016;126:4727–34.
Mantzoros CS. The role of leptin in human obesity and disease: a review of current evidence. Ann Intern Med. 1999;130:671–80.
Liu J, Guo M, Lu XY. Leptin/LepRb in the ventral tegmental area mediates anxiety-related behaviors. Int J Neuropsychopharmacol. 2015;19:pyv115.
Guo M, Lu Y, Garza JC, Li Y, Chua SC, Zhang W, et al. Forebrain glutamatergic neurons mediate leptin action on depression-like behaviors and synaptic depression. Transl Psychiatry. 2012;2:e83.
Wang X, Zhang D, Lu XY. Dentate gyrus-CA3 glutamate release/NMDA transmission mediates behavioral despair and antidepressant-like responses to leptin. Mol Psychiatry. 2015;20:509–19.
Lu XY, Kim CS, Frazer A, Zhang W. Leptin: a potential novel antidepressant. Proc Natl Acad Sci USA. 2006;103:1593–8.
Chuang JC, Krishnan V, Yu HG, Mason B, Cui H, Wilkinson MB, et al. A beta3-adrenergic-leptin-melanocortin circuit regulates behavioral and metabolic changes induced by chronic stress. Biol Psychiatry. 2010;67:1075–82.
Finger BC, Dinan TG, Cryan JF. High-fat diet selectively protects against the effects of chronic social stress in the mouse. Neuroscience. 2011;192:351–60.
Garza JC, Guo M, Zhang W, Lu XY. Leptin restores adult hippocampal neurogenesis in a chronic unpredictable stress model of depression and reverses glucocorticoid-induced inhibition of GSK-3beta/beta-catenin signaling. Mol Psychiatry. 2012;17:790–808.
Liu J, Garza JC, Bronner J, Kim CS, Zhang W, Lu XY. Acute administration of leptin produces anxiolytic-like effects: a comparison with fluoxetine. Psychopharmacology. 2010;207:535–45.
Yamada N, Katsuura G, Ochi Y, Ebihara K, Kusakabe T, Hosoda K, et al. Impaired CNS leptin action is implicated in depression associated with obesity. Endocrinology. 2011;152:2634–43.
Lawson EA, Miller KK, Blum JI, Meenaghan E, Misra M, Eddy KT, et al. Leptin levels are associated with decreased depressive symptoms in women across the weight spectrum, independent of body fat. Clin Endocrinol. 2012;76:520–5.
Guo M, Huang TY, Garza JC, Chua SC, Lu XY. Selective deletion of leptin receptors in adult hippocampus induces depression-related behaviours. Int J Neuropsychopharmacol. 2013;16:857–67.
Duman RS, Deyama S, Fogaca MV. Role of BDNF in the pathophysiology and treatment of depression: activity-dependent effects distinguish rapid-acting antidepressants. Eur J Neurosci. 2019. https://doi.org/10.1111/ejn.14630. [Online ahead of print].
Nibuya M, Morinobu S, Duman RS. Regulation of BDNF and trkB mRNA in rat brain by chronic electroconvulsive seizure and antidepressant drug treatments. J Neurosci. 1995;15:7539–47.
Russo-Neustadt AA, Beard RC, Huang YM, Cotman CW. Physical activity and antidepressant treatment potentiate the expression of specific brain-derived neurotrophic factor transcripts in the rat hippocampus. Neuroscience. 2000;101:305–12.
Chen B, Dowlatshahi D, MacQueen GM, Wang JF, Young LT. Increased hippocampal BDNF immunoreactivity in subjects treated with antidepressant medication. Biol Psychiatry. 2001;50:260–5.
Saarelainen T, Hendolin P, Lucas G, Koponen E, Sairanen M, MacDonald E, et al. Activation of the TrkB neurotrophin receptor is induced by antidepressant drugs and is required for antidepressant-induced behavioral effects. J Neurosci. 2003;23:349–57.
Siuciak JA, Lewis DR, Wiegand SJ, Lindsay RM. Antidepressant-like effect of brain-derived neurotrophic factor (BDNF). Pharmacol Biochem Behav. 1997;56:131–7.
Shirayama Y, Chen AC, Nakagawa S, Russell DS, Duman RS. Brain-derived neurotrophic factor produces antidepressant effects in behavioral models of depression. J Neurosci. 2002;22:3251–61.
Autry AE, Adachi M, Nosyreva E, Na ES, Los MF, Cheng PF, et al. NMDA receptor blockade at rest triggers rapid behavioural antidepressant responses. Nature. 2011;475:91–5.
Liu RJ, Lee FS, Li XY, Bambico F, Duman RS, Aghajanian GK. Brain-derived neurotrophic factor Val66Met allele impairs basal and ketamine-stimulated synaptogenesis in prefrontal cortex. Biol Psychiatry. 2012;71:996–1005.
Timmusk T, Palm K, Metsis M, Reintam T, Paalme V, Saarma M, et al. Multiple promoters direct tissue-specific expression of the rat BDNF gene. Neuron. 1993;10:475–89.
Aid T, Kazantseva A, Piirsoo M, Palm K, Timmusk T. Mouse and rat BDNF gene structure and expression revisited. J Neurosci Res. 2007;85:525–35.
Lauterborn JC, Rivera S, Stinis CT, Hayes VY, Isackson PJ, Gall CM. Differential effects of protein synthesis inhibition on the activity-dependent expression of BDNF transcripts: evidence for immediate-early gene responses from specific promoters. J Neurosci. 1996;16:7428–36.
Nanda S, Mack KJ. Multiple promoters direct stimulus and temporal specific expression of brain-derived neurotrophic factor in the somatosensory cortex. Brain Res Mol Brain Res. 1998;62:216–9.
Yang J, Siao CJ, Nagappan G, Marinic T, Jing D, McGrath K, et al. Neuronal release of proBDNF. Nat Neurosci. 2009;12:113–5.
Hong EJ, McCord AE, Greenberg ME. A biological function for the neuronal activity-dependent component of Bdnf transcription in the development of cortical inhibition. Neuron. 2008;60:610–24.
West AE, Pruunsild P, Timmusk T. Neurotrophins: transcription and translation. Handb Exp Pharmacol. 2014;220:67–100.
Tuvikene J, Pruunsild P, Orav E, Esvald EE, Timmusk T. AP-1 transcription factors mediate BDNF-positive feedback loop in cortical neurons. J Neurosci. 2016;36:1290–305.
Smith MA, Makino S, Kvetnansky R, Post RM. Stress and glucocorticoids affect the expression of brain-derived neurotrophic factor and neurotrophin-3 mRNAs in the hippocampus. J Neurosci. 1995;15:1768–77.
Smith MA, Cizza G. Stress-induced changes in brain-derived neurotrophic factor expression are attenuated in aged Fischer 344/N rats. Neurobiol Aging. 1996;17:859–64.
Ueyama T, Kawai Y, Nemoto K, Sekimoto M, Tone S, Senba E. Immobilization stress reduced the expression of neurotrophins and their receptors in the rat brain. Neurosci Res. 1997;28:103–10.
Marmigere F, Givalois L, Rage F, Arancibia S, Tapia-Arancibia L. Rapid induction of BDNF expression in the hippocampus during immobilization stress challenge in adult rats. Hippocampus. 2003;13:646–55.
Dias BG, Banerjee SB, Duman RS, Vaidya VA. Differential regulation of brain derived neurotrophic factor transcripts by antidepressant treatments in the adult rat brain. Neuropharmacology. 2003;45:553–63.
Oliff HS, Berchtold NC, Isackson P, Cotman CW. Exercise-induced regulation of brain-derived neurotrophic factor (BDNF) transcripts in the rat hippocampus. Brain Res Mol Brain Res. 1998;61:147–53.
Russo-Neustadt AA, Alejandre H, Garcia C, Ivy AS, Chen MJ. Hippocampal brain-derived neurotrophic factor expression following treatment with reboxetine, citalopram, and physical exercise. Neuropsychopharmacology. 2004;29:2189–99.
Martinowich K, Manji H, Lu B. New insights into BDNF function in depression and anxiety. Nat Neurosci. 2007;10:1089–93.
DeFalco J, Tomishima M, Liu H, Zhao C, Cai X, Marth JD, et al. Virus-assisted mapping of neural inputs to a feeding center in the hypothalamus. Science. 2001;291:2608–13.
Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–40.
Rios M, Fan G, Fekete C, Kelly J, Bates B, Kuehn R, et al. Conditional deletion of brain-derived neurotrophic factor in the postnatal brain leads to obesity and hyperactivity. Mol Endocrinol. 2001;15:1748–57.
Gorski JA, Zeiler SR, Tamowski S, Jones KR. Brain-derived neurotrophic factor is required for the maintenance of cortical dendrites. J Neurosci. 2003;23:6856–65.
Lei Y, Wang J, Wang D, Li C, Liu B, Fang X, et al. SIRT1 in forebrain excitatory neurons produces sexually dimorphic effects on depression-related behaviors and modulates neuronal excitability and synaptic transmission in the medial prefrontal cortex. Mol Psychiatry. 2020;25:1094–111.
Carrier N, Wang X, Sun L, Lu XY. Sex-specific and estrous cycle-dependent antidepressant-like effects and hippocampal Akt signaling of leptin. Endocrinology. 2015;156:3695–705.
Chua SC Jr., Chung WK, Wu-Peng XS, Zhang Y, Liu SM, Tartaglia L, et al. Phenotypes of mouse diabetes and rat fatty due to mutations in the OB (leptin) receptor. Science. 1996;271:994–6.
Lee GH, Proenca R, Montez JM, Carroll KM, Darvishzadeh JG, Lee JI, et al. Abnormal splicing of the leptin receptor in diabetic mice. Nature. 1996;379:632–5.
Leinninger GM, Jo YH, Leshan RL, Louis GW, Yang H, Barrera JG, et al. Leptin acts via leptin receptor-expressing lateral hypothalamic neurons to modulate the mesolimbic dopamine system and suppress feeding. Cell Metab. 2009;10:89–98.
Liao GY, An JJ, Gharami K, Waterhouse EG, Vanevski F, Jones KR. et al. Dendritically targeted Bdnf mRNA is essential for energy balance and response to leptin. Nat Med. 2012;18:564–71.
Wu MV, Sahay A, Duman RS, Hen R. Functional differentiation of adult-born neurons along the septotemporal axis of the dentate gyrus. Cold Spring Harb Perspect Biol. 2015;7:a018978.
Klein R, Nanduri V, Jing SA, Lamballe F, Tapley P, Bryant S, et al. The trkB tyrosine protein kinase is a receptor for brain-derived neurotrophic factor and neurotrophin-3. Cell. 1991;66:395–403.
Notaras M, van den Buuse M. Neurobiology of BDNF in fear memory, sensitivity to stress, and stress-related disorders. Mol Psychiatry. 2020;25:2251–74.
Atwal JK, Massie B, Miller FD, Kaplan DR. The TrkB-Shc site signals neuronal survival and local axon growth via MEK and P13-kinase. Neuron. 2000;27:265–77.
Chen H, Charlat O, Tartaglia LA, Woolf EA, Weng X, Ellis SJ, et al. Evidence that the diabetes gene encodes the leptin receptor: identification of a mutation in the leptin receptor gene in db/db mice. Cell. 1996;84:491–5.
Fungfuang W, Nakada T, Nakao N, Terada M, Yokosuka M, Gizurarson S, et al. Serum leptin concentrations, leptin mRNA expression, and food intake during the estrous cycle in rats. Lab Anim Res. 2013;29:1–6.
Donato J Jr, Cravo RM, Frazao R, Elias CF. Hypothalamic sites of leptin action linking metabolism and reproduction. Neuroendocrinology. 2011;93:9–18.
Allen SJ, Garcia-Galiano D, Borges BC, Burger LL, Boehm U, Elias CF. Leptin receptor null mice with reexpression of LepR in GnRHR expressing cells display elevated FSH levels but remain in a prepubertal state. Am J Physiol Regul Integr Comp Physiol. 2016;310:R1258–1266.
Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, et al. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–9.
Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, et al. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–3.
Park HK, Ahima RS. Leptin signaling. F1000Prime Rep. 2014;6:73.
Huang WC, Chen CC. Akt phosphorylation of p300 at Ser-1834 is essential for its histone acetyltransferase and transcriptional activity. Mol Cell Biol. 2005;25:6592–602.
Bowers EM, Yan G, Mukherjee C, Orry A, Wang L, Holbert MA, et al. Virtual ligand screening of the p300/CBP histone acetyltransferase: identification of a selective small molecule inhibitor. Chem Biol. 2010;17:471–82.
Tang Z, Chen WY, Shimada M, Nguyen UT, Kim J, Sun XJ, et al. SET1 and p300 act synergistically, through coupled histone modifications, in transcriptional activation by p53. Cell. 2013;154:297–310.
Leuner B, Gould E. Structural plasticity and hippocampal function. Annu Rev Psychol. 2010;61:111–40.
Cooper C, Moon HY, van Praag H. On the run for hippocampal plasticity. Cold Spring Harb Perspect Med. 2018;8:a029736.
Branchi I, Francia N, Alleva E. Epigenetic control of neurobehavioural plasticity: the role of neurotrophins. Behav Pharmacol. 2004;15:353–62.
Lu B, Gottschalk W. Modulation of hippocampal synaptic transmission and plasticity by neurotrophins. Prog Brain Res. 2000;128:231–41.
Groves JO. Is it time to reassess the BDNF hypothesis of depression? Mol Psychiatry. 2007;12:1079–88.
Dwivedi Y, Rizavi HS, Conley RR, Roberts RC, Tamminga CA, Pandey GN. Altered gene expression of brain-derived neurotrophic factor and receptor tyrosine kinase B in postmortem brain of suicide subjects. Arch Gen Psychiatry. 2003;60:804–15.
Karege F, Vaudan G, Schwald M, Perroud N, La Harpe R. Neurotrophin levels in postmortem brains of suicide victims and the effects of antemortem diagnosis and psychotropic drugs. Brain Res Mol Brain Res. 2005;136:29–37.
Aydemir O, Deveci A, Taneli F. The effect of chronic antidepressant treatment on serum brain-derived neurotrophic factor levels in depressed patients: a preliminary study. Prog Neuropsychopharmacol Biol Psychiatry. 2005;29:261–5.
Duman RS. Role of neurotrophic factors in the etiology and treatment of mood disorders. Neuromol Med. 2004;5:11–25.
Govindarajan A, Rao BS, Nair D, Trinh M, Mawjee N, Tonegawa S, et al. Transgenic brain-derived neurotrophic factor expression causes both anxiogenic and antidepressant effects. Proc Natl Acad Sci USA. 2006;103:13208–13.
Monteggia LM, Barrot M, Powell CM, Berton O, Galanis V, Gemelli T, et al. Essential role of brain-derived neurotrophic factor in adult hippocampal function. Proc Natl Acad Sci USA. 2004;101:10827–32.
Adachi M, Barrot M, Autry AE, Theobald D, Monteggia LM. Selective loss of brain-derived neurotrophic factor in the dentate gyrus attenuates antidepressant efficacy. Biol Psychiatry. 2008;63:642–9.
Monteggia LM, Luikart B, Barrot M, Theobold D, Malkovska I, Nef S, et al. Brain-derived neurotrophic factor conditional knockouts show gender differences in depression-related behaviors. Biol Psychiatry. 2007;61:187–97.
Bjorkholm C, Monteggia LM. BDNF—a key transducer of antidepressant effects. Neuropharmacology. 2016;102:72–9.
Maynard KR, Hill JL, Calcaterra NE, Palko ME, Kardian A, Paredes D, et al. Functional role of BDNF production from unique promoters in aggression and serotonin signaling. Neuropsychopharmacology. 2016;41:1943–55.
Musazzi L, Rimland JM, Ieraci A, Racagni G, Domenici E, Popoli M. Pharmacological characterization of BDNF promoters I, II and IV reveals that serotonin and norepinephrine input is sufficient for transcription activation. Int J Neuropsychopharmacol. 2014;17:779–91.
Alme MN, Wibrand K, Dagestad G, Bramham CR. Chronic fluoxetine treatment induces brain region-specific upregulation of genes associated with BDNF-induced long-term potentiation. Neural Plast. 2007;2007:26496.
Dwivedi Y, Rizavi HS, Pandey GN. Antidepressants reverse corticosterone-mediated decrease in brain-derived neurotrophic factor expression: differential regulation of specific exons by antidepressants and corticosterone. Neuroscience. 2006;139:1017–29.
Badeaux AI, Shi Y. Emerging roles for chromatin as a signal integration and storage platform. Nat Rev Mol Cell Biol. 2013;14:211–24.
Cha TL, Zhou BP, Xia W, Wu Y, Yang CC, Chen CT, et al. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science. 2005;310:306–10.
Karmodiya K, Krebs AR, Oulad-Abdelghani M, Kimura H, Tora L. H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genom. 2012;13:424.
Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37.
Gong F, Miller KM. Histone methylation and the DNA damage response. Mutat Res. 2019;780:37–47.
Garza JC, Guo M, Zhang W, Lu XY. Leptin increases adult hippocampal neurogenesis in vivo and in vitro. J Biol Chem. 2008;283:18238–47.
McGregor G, Harvey J. Regulation of hippocampal synaptic function by the metabolic hormone, leptin: implications for health and neurodegenerative disease. Front Cell Neurosci. 2018;12:340.
Acknowledgements
This work was supported by NSFSP No. ZR2014HQ080 (to CL) and NIH grants MH100583, MH119456, AG062166 and AG064895 (to X-YL).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, C., Meng, F., Lei, Y. et al. Leptin regulates exon-specific transcription of the Bdnf gene via epigenetic modifications mediated by an AKT/p300 HAT cascade. Mol Psychiatry 26, 3701–3722 (2021). https://doi.org/10.1038/s41380-020-00922-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-020-00922-0
This article is cited by
-
Role of Hippocampal miR-132-3p in Modifying the Function of Protein Phosphatase Mg2+/Mn2+-dependent 1 F in Depression
Neurochemical Research (2023)
-
Leptin enhances social motivation and reverses chronic unpredictable stress-induced social anhedonia during adolescence
Molecular Psychiatry (2022)
-
Gm364 coordinates MIB2/DLL3/Notch2 to regulate female fertility through AKT activation
Cell Death & Differentiation (2022)
-
PPM1F in Dentate Gyrus Modulates Anxiety-Related Behaviors by Regulating BDNF Expression via AKT/JNK/p-H3S10 Pathway
Molecular Neurobiology (2021)