Abstract
ADP-ribosylation (ADPR) is a posttranslational modification whose importance in oncology keeps increasing due to frequent use of PARP inhibitors (PARPi) to treat different tumor types. Due to the lack of suitable tools to analyze cellular ADPR levels, ADPR’s significance for cancer progression and patient outcome is unclear. In this study, we assessed ADPR levels by immunohistochemistry using a newly developed anti-ADP-ribose (ADPr) antibody, which is able to detect both mono- and poly-ADPR. Tissue microarrays containing brain (n = 103), breast (n = 1108), colon (n = 236), lung (n = 138), ovarian (n = 142), and prostate (n = 328) cancers were used to correlate ADPR staining intensities to clinico-pathological data, including patient overall survival (OS), tumor grade, tumor stage (pT), lymph node status (pN), and the presence of distant metastasis (pM). While nuclear ADPR was detected only in a minority of the samples, cytoplasmic ADPR (cyADPR) staining was observed in most tumor types. Strong cyADPR intensities were significantly associated with better overall survival in invasive ductal breast cancer (p < 0.0001), invasive lobular breast cancer (p < 0.005), and high grade serous ovarian cancer patients (p < 0.01). Furthermore, stronger cytoplasmic ADPR levels significantly correlated with early tumor stage in colorectal and in invasive ductal breast adenocarcinoma (p < 0.0001 and p < 0.01, respectively) and with the absence of regional lymph node metastasis in colorectal adenocarcinoma (p < 0.05). No correlation to cyADPR was found for prostate and lung cancer or brain tumors. In conclusion, our new anti-ADP-ribose antibody revealed heterogeneous ADPR staining patterns with predominant cytoplasmic ADPR staining in most tumor types. Different cyADPR staining patterns could help to better understand variable response rates to PARP inhibitors in the future.
Similar content being viewed by others
Introduction
Some of the most recently approved drugs for the treatment of breast [1, 2] and ovarian [3, 4] cancer are poly-(ADP-ribose) polymerases (PARP) inhibitors (PARPi). Two breakthrough studies in preclinical models demonstrated the effectiveness of PARPi in the treatment of these cancers by showing synthetic lethality between the PARPi and the mutations in BRCA1/2 encoding components of homologous recombination-mediated DNA repair (HRR) pathways [5, 6]. Recently, the beneficial effect of Olaparib (PARPi) in treating metastatic castration-resistant prostate cancer patients was reported in a profound clinical study [7]. The use of PARPi is also considered in clinical trials for the treatment of several other tumor types, including lung cancer [8], pancreatic cancer [9], melanoma [10], glioblastoma [11], colorectal cancer [12,13,14], and renal cell carcinoma [15, 16], either alone or in combination with other drugs, and sometimes even independently of the BRCA status [17]. However, besides these encouraging results, the use of PARPi remains clinically relegated to few cancer types and their administration relays mainly on patient stratification based on “BRCAness” [18], obtained by gene mutation analysis of BRCA1/2 and its modulators, without considering the actual PARP activities. Mechanistically, currently approved PARP inhibitors inhibit the activity of some ADP-ribosyl transferases (ARTs), also known as PARPs [19]. ARTs and their counterparts, ADPr-glycohydrolases, modulate the levels of ADP-ribosylation (ADPR), a reversible posttranslational modification (PTM) [20], by transferring ADP-ribose (ADPr) moieties from Nicotinamide Adenine Dinucleotide (NAD+) to specific amino acid acceptor sites on target proteins [21]. ADPR exists in two forms, namely: mono-ADP-ribosylation (MARylation), which consists of only one ADPr monomer linked to the target protein, and poly-ADP-ribosylation (PARylation), a chain(s) of ADPr moieties extending from the MARylated form of the target protein [20]. PARylation was intensively studied in the last decades and was shown to be involved in transcription control, protein degradation, chromatin organization, and DNA repair [6, 22]. MARylation has been reported in cell-based assays involved in tumorigenesis since ARTs known to only MARylate proteins also regulate several cellular processes critical for cancer cells, such as cell proliferation, apoptosis, growth, and metabolism [23]. Moreover, some of these ARTs were also reported to modulate the activity of proteins involved in molecular pathways that are frequently dysregulated in cancer cells, such as c-Myc and NF-κB [24, 25].
So far, MARylation has not been investigated by immunohistochemistry on patient histological sections due to the missing tools. The main reason for this lack can be ascribed to the difficulties encountered in the chemical synthesis of ADP-ribosylated peptides required for the host immunization and the generation of antibodies. Indeed, the chemical nature of the linkage and the variety of modified acceptor amino acids requires long, complicated synthesis processes that yield only low amounts of peptides [26, 27]. Therefore, so far, only few studies have investigated endogenous ADPR (MAR and PAR) as a potential biomarker in oncology [23, 28, 29]. One of these [29] recently provided evidence by immunoblotting that PARP1/ARTD1-dependent ADPR levels correlate with clinical outcome in high grade serous ovarian cancer patients, whereas the other studies mainly relayed on mass spectrometry-based approaches and their results about the role of ADPR in cancer are partially contradicting [23, 28]. However, a comprehensive cancer study about ADPR (MAR and PAR) that particularly focuses on the analysis of cellular staining patterns in different human tumors is still missing.
Following an established methodology [30], we recently successfully synthesized ADP-ribosylated peptides and produced a polyclonal antibody that detects both MARylated and PARylated proteins in Westernblot and Immunfluorescence independently of the binding site and without cross-reacting with other posttranslational modifications [31, 32]. By immunohistochemical staining of various tissue microarrays (TMAs) using this new anti-ADPr antibody, we investigated the cellular staining patterns of ADPR in different cancer types, as well as their relevance for tumor progression.
Material and methods
Cell culture and pellets
HeLa cells (Kyoto, ATCC, CCL-2) were cultured in Dulbecco’s modified Eagle’s medium (DMEM), supplemented with 10% fetal calf serum (FCS) and 1% penicillin/streptavidin at 37 °C with 5% CO2 until 80% confluency. Nuclear ADP-ribosylation was induced in HeLa cells by treatment with 1 mM H2O2 in PBS containing 1 mM MgCl2 for 10 min. Cell pellets were obtained after coincubation with human plasma in presence of thrombin and consequently fixed with 4% formalin, dehydrated, and paraffinized.
Patient tissue samples and tissue microarray construction
Formalin fixed and paraffin embedded tumor samples were retrieved from the archives of the Department of Pathology and Molecular Pathology, University Hospital Zurich (Zurich, Switzerland) between the years 1993 and 2013, thus the patients were not treated with approved PARP inhibitors. For each tumor, one representative tumor tissue block was re-evaluated using hematoxylin and eosin-stained sections. Only those cases with representative tumor regions that contained at least 70% tumor cells were selected for tissue microarray (TMA) construction. All tumors were reviewed by pathologists of the Department of Pathology and Molecular Pathology specialized in their field. Classification, grading, and staging were performed according to current TNM and WHO classification. The TMAs contained 1108 breast carcinomas (three TMAs 336 + 344 + 428) [33], 142 ovarian carcinomas [34], 328 prostate carcinomas [35], 138 lung carcinomas [36], 236 colorectal carcinomas (two TMAs 101 + 135) [37], and 103 brain tumors [38]. The TMAs were constructed as described [39] and each punch had a diameter of 0.6 mm. This study was approved by the local commission of ethics (BASEC-Nr_2016-00811).
Immunohistochemistry (IHC) staining
In total, 2.5 μm sections of TMAs and paraffinized cell pellets were placed on glass slides and immunohistochemically stained using Ultra Discovery (Ventana, Roche Diagnostics, Rotkreuz, Switzerland). TMA sections were pretreated with Tris-EDTA-Borate Buffer of pH 9 at 95 °C for 60 min (CC1 standard protocol, Ventana) and incubated with anti-ADPr antibody [31, 32] (rabbit, diluted 1:500 in Bond medium) for 44 min at 36 °C (Discovery Ultra, Ventana). ADPr was made visible using the UltraMap-Rabbit DAB detection kit (Ventana). Nuclear counterstaining was performed with hematoxylin. The polyclonal anti-ADPr antibody is not commercialized and it is available upon request if used only for academic purposes.
Scoring of the IHC staining
For staining evaluation, stained slides were scanned using a NanoZoomer (Hamamatsu Photonics, Shizuoka, Japan). The images were analyzed with ObjectiveViewTM software. Tumors were scored as strong (+3), moderate (+2), or weak/negative (+1) for cytoplasmic ADPR (cyADPR) in the tumor cells. ADPR in the epithelium of normal tissue samples and nuclear ADPR signal in cancer biopsies among the TMAs were considered as positive reference for ADPR staining. The scores were blindly double-checked by an independent pathologist.
Statistical analysis
The software Graphpad Prism 8 was used to run statistical analysis. The patient survival was reported with Kaplan–Meier chart and assessed with Mantel–Cox statistical analysis. Correlation of ADPR scores with pathological parameters was assessed by contingency analysis, chi-square test with a confidence interval (CI) of 95%. Univariate and multivariate analysis were performed with R Studio software. Cox proportional model was used for the statistical analysis as described [40]. Differences were considered significant when p value < 0.05.
Immunofluorescent (IF) staining
In total, 2.5 μm sections of TMAs and paraffinized cell pellets were placed on glass slides and dried for 10 min at 60 °C, dewaxed for 10 min in HistoChoice Clearing Agent (Sigma, H2779), and rehydrated by consecutive washes in ethanol: 100%, 95%, 70%, and 40% v/v for 30 sec each. Heat-induced antigen retrieval was performed at 95 °C for 40 min with Dako Target Retrieval Solution of pH 9 (Dako, S2367). After cooling at room temperature, the slides were incubated 2.5% BSA (Fluka Analytical, 05488) in TBST 0.1% for 1 hour at room temperature. Anti-ADPr [31, 32] (rabbit, 1:500 dilution) and anti-ATP5a antibody (Abcam, mouse, 1:250) diluted in the blocking solution were incubated overnight at 4 °C. Secondary antibody Alexa Fluor 488 AffiniPure goat anti-mouse (Jackson ImmunoResearch, 115–545–003, 1:500 dilution) and Cy3 AffiniPure goat anti-rabbit IgG (Jackson ImmunoResearch, 111–165–144, 1:250 dilution) were incubated for 2 hours at room temperature. Nuclear counterstaining was performed DAPI (Biolegend, 422801, 1:10,000 dilution). Microscope cover glasses (Menzel–Glaser, 7001023) were mounted with Mowiol mounting medium.
Fluorescent and confocal microscopy
Fluorescent microscopy images were acquired with Leica CTR6000, 63x magnification, 250 and 400 ms exposure time (for DAPI and ADPR respectively), 100% FIM. Slides stained without the primary antibody were used as technical negative control. Focal plan was established on DAPI signal.
Confocal microscopy images were acquired with the automated CLSM––Leica SP8 upright confocal laser scanning microscope equipped with four solid state diode lasers (405, 488, 522, and 638 nm), using an HCX PL APO CS2 63x immersion oil objective present at the University of Zurich Center for Microscope and Image Analysis (ZMB). Nuclear focus was used to establish the focal plane.
The images were analyzed with the Leica software.
Results
Immunostaining with the anti-ADPr antibody is repeatable and detects ADP-ribosylation in different cellular compartments
To investigate the possible implication of ADP-ribosylation (both MAR- and PARylation) in cancer we performed a comprehensive study by immunohistochemically staining TMAs with a recently developed anti-ADP-ribose (ADPr) antibody that recognizes both MAR- and PARylation [31, 32]. To first characterize the ADPR staining patterns of the antibody for immunofluorescence (IF) and immunocytochemistry (ICC) we stained HeLa cell pellets either untreated or incubated with hydrogen peroxide. Both methodologies revealed that untreated cells were characterized by a diffuse cytoplasmic ADPR signal, whereas the H2O2 treatment, known to induce nuclear ADP-ribosylation [32], led to the detection of strong nuclear ADPR signal (Suppl. Figs 1, 2). Moreover, the IF and ICC stainings of the cell pellets with the anti-ADPr antibody performed in different days were repeatable and showed comparable signal intensities and distributions (Suppl. Figs 1, 2). Next, we optimized the immunohistochemical staining on several tumors included in a test tissue microarray (TMA). Also here, the ADPR stainings were consistent in terms of signal intensity and distribution (i.e., pattern) in different repetitions (Suppl. Fig. 3).
Differential ADP-ribosylation is predominantly observed in the cell cytoplasm
We started our comprehensive tumor ADPR study by immunohistochemically staining three breast cancer TMAs using the anti-ADPr antibody according to our optimized staining protocol. Overall analysis of the signal localization and intensity revealed that a very weak homogenous nuclear ADPR signal (nuADPR) staining was observed only in rare cases of healthy tissues and for only a few breast cancer biopsies (12 out of 150) (Supplementary Fig. 4). Furthermore, intra-nuclear foci were observed in many cancer cells and in nearly all initially analyzed biopsies, but the weak ADPR signal staining did not allow to discriminate and score the signal intensities (data not shown). On the other hand, cytoplasmic ADPR (cyADPR) signal was observed in all the biopsies with different intensities (Fig. 1A). Thus, we decided to focus our study exclusively on the cytoplasmic ADPR (cyADPR) signal intensities.
A IHC staining of a breast cancer TMA using the anti-ADPr antibody. cyADPR staining intensity scores: strong (red, 3), moderate (yellow, 2), and weak/negative (blue, 1). On the top, selected TMA cores (scale bar 100 µm, 10x), on the bottom, ROIs (scale bar 20 µm, 40x). Kaplan–Meier survival plot of (B) invasive ductal breast cancer (Mantel–Cox test, p value < 0.0001, N = 643) and (C) invasive lobular breast cancer (Mantel–Cox test, p value = 0.0017, N = 116) with patients stratified based on the cyADPR signal intensity scores. Contingency analysis to assess the association between cyADPR scores and the tumor stage (pT 1–4), the tumor grade (1–3), regional lymph node metastasis (pN 0–2), and the presence of distant metastasis (pM 0–1) in invasive ductal breast cancer (D, F, H, J) and in invasive lobular breast cancer (E, G, I, K).
Strong cytoplasmic ADPR correlates with better outcome in breast cancer patients
Cytoplasmic ADPR signal intensities in breast cancer biopsies were scored using three categories (3: strong, 2: moderate, and 1: weak/negative) and subsequently correlated to patient clinical data including overall survival (OS), nuclear differentiation grade, tumor stage (pT), regional lymph node metastasis (pN), and distant metastasis (pM). In invasive ductal breast cancer (IDC), strong cyADPR intensity significantly correlated with better patient overall survival, whereas a weak/negative cyADPR signal intensity was associated with a poor patient prognosis (n = 643, p < 0.0001, Fig. 1B). Also, in invasive lobular breast cancer (ILC) biopsies, similarly to the results mentioned before, strong cyADPR was significantly associated with better patient outcome (n = 116, p = 0.0017, Fig. 1C).
In IDC, we observed that pT1 tumors displayed in 50% of the cases a strong cyADPR intensity, whereas in pT3 and pT4 cases strong cyADPR signal intensity significantly dropped to 28% and 33%, respectively (n = 705, p = 0.0097 and 0.0076, Fig. 1D). In ILC, we observed a gradual increase of weak/negative cyADPR intensity scores incidence from pT1 (2%) to pT4 (21%) but this difference was not statistically significant (n = 130, Fig. 1E). No significant correlation was obtained between cyADPR intensities and nuclear differentiation grade in both IDC (n = 703, Fig. 1F) and ILC (n = 118, Fig. 1G). In contrast, an interesting tendency although not statistically significant between decreased cyADPR levels and the presence of metastasis in regional lymph node (pN1/2) was noticed for IDC (n = 459, Fig. 1H) but not for ILC (n = 74, Fig. 1I). No correlation was obtained for cyADPR intensities and the presence of distant metastasis (pM) neither in IDC (n = 42, Fig. 1J) nor in ILC (n = 22, Fig. 1K).
Multivariate analysis assessed with the Cox multivariate regression test (test likelihood ratio p = 8e–10) revealed that in IDC (n = 411, number of events = 127) cyADPR levels (β = −0.25, CI = 95%, HR = 0.78, p = 0.059) were not independent from tumor stage (β = 0.43, CI = 95%, HR = 1.53, p < 0.0001), nodular stage (β = 0.26, CI = 95%, HR = 1.30, p = 0.0348), and tumor grade (β = 0.49, CI = 95%, HR = 1.63, p = 0.0018) (Table 1). In ILC, due to the lower number of cases (n = 51) and events (n = 15) the multivariate analysis assessed with the Cox multivariate regression test (test likelihood ratio p = 0.002), did not yield significant outcome as obtained in IDC (Table 1). The data, however, indicate a trend, which is similar to that seen in IDC.
Strong cytoplasmic ADPR correlates with a better prognosis in high grade serous ovarian cancer
To assess whether cyADPR levels have prognostic impact in ovarian cancer, we analyzed 142 ovarian tumors for cyADPR signal intensities (Fig. 2A). By distinguishing between the ovarian cancer subtypes high grade serous (n = 68), clear cell (n = 26), endometrioid (n = 29), and mucinous (n = 14), we observed a significant correlation between high cyADPR levels and prolonged patient survival for high grade serous OC, the most abundant ovarian cancer subtype in the TMA (p = 0.0088, Fig. 2B). We also observed reduced cyADPR intensity in advanced tumor stage (pT) however, this tendency was not statistically significant (n = 68, Fig. 2C).
A IHC staining of an ovarian cancer TMA using the anti-ADPr antibody. Staining intensity scores: strong (red, 3), moderate (yellow, 2), and weak/negative (blue, 1). On the top, selected TMA cores (scale bar 100 µm, 10x), on the bottom, ROIs (scale bar 20 µm, 40x). B Kaplan–Meier survival plot of high grade serous ovarian cancer (Mantel–Cox test, p value = 0.0088, N = 68) with patients stratified based on the cyADPR signal intensity scores. C Contingency analysis to assess the association between cyADPR scores and the tumor stage (pT 1–4) in high grade serous ovarian cancer.
Cytoplasmic ADPR signal intensities do not associate with prostate, lung, and brain cancer patient prognosis
Recent studies indicate the importance of ADPR in prostate [41, 42], lung [43, 44], and brain cancer [45]. We therefore analyzed cyADPR intensities of these cancer types using the weak, moderate, and strong cyADPR intensity scoring. Although staining differences could be observed, in all three tested cancer types, there were no significant correlations between cyADPR signal intensities and overall survival (OS), tumor stage (pT), differentiation grade or the metastasis presence (pN and pM), when data were available (Supplementary Figs 5, 6 and 7). Interestingly, lung adenocarcinomas with weak cyADPR signal intensities showed a trend to shorter patient survival, whereas this trend was not observed in lung squamous cell cancer.
Weak cytoplasmic ADPR signal intensity correlates with late tumor stages and with lymph node metastasis in colon adenocarcinoma
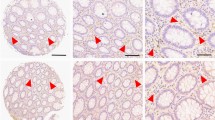
Recently an association between ADPR and colon cancer development was suggested [46, 47]. We investigated cyADPR prognostic relevance in colon adenocarcinoma by analyzing cyADPR signal intensities in 256 biopsies allocated to two TMAs (Fig. 3A). Since patient survival data were lacking for the majority of the cases for this tumor type, it was only possible to associate the cyADPR scores to the classical prognostic markers tumor stage (pT), grade and metastasis (pN and pM). Interestingly, colon adenocarcinomas in pT1 were characterized in 85% by a strong cyADPR staining, whereas pT3 and pT4 tumors were characterized in 43% and 25%, respectively, by a strong cyADPR staining intensity. Together these data indicated a statistically significant decrease of cyADPR staining during tumor progression (Fig. 3B). No significant correlation was obtained between cyADPR intensities and nuclear differentiation grade (n = 220, Fig. 3C). Next, cyADPR signal intensity was correlated with the presence of metastasis. Half of the tumors lacking regional lymph node metastasis (pN0) had strong and only 6% weak/negative cyADPR signal intensity, whereas 34% of tumors with regional lymph node metastasis (pN ≥ 1) presented with a strong and 17% with a weak/negative cyADPR staining (p = 0.0116, n = 227, Fig. 3D). No significant correlation was obtained between cyADPR intensities and the presence of distant metastasis (pM) (n = 46, Fig. 3E).
A IHC staining of a colon cancer TMA using the anti-ADPr antibody. Staining intensity scores: strong (red, 3), moderate (yellow, 2), and weak/negative (blue, 1). On the top, selected TMA cores (scale bar 100 µm, 10x), on the bottom, ROIs (scale bar 20 µm, 40x). Contingency analyses to assess the association between cyADPR scores and (B) the tumor stage (pT 1–4), (C) the tumor grade (1–3), (D) regional lymph node metastasis (pN 0–2), and (E) the presence of distant metastasis (pM 0–1).
Mitochondrial ADP-ribosylome is the major source of cytoplasmic ADPR in selected tumor types
To better identify the cellular location of the cyADPR, we colocalized by confocal immunofluorescent microscopy the ADPR signal with an organelle-specific marker in different patient biopsies. Since recent studies showed mitochondria ADPR as major source of cellular ADP-ribosylome [48, 49], we co-stained breast, colon, lung, and ovarian cancer biopsies using the anti-ADPr antibody and an antibody specific for the mitochondrial marker ATP5a. We observed that the ADPR signal was frequently but not exclusively overlapping with the mitochondrial marker ATP5a, indicating that mitochondrial ADP-ribosylated proteins is one of the major sources of cytoplasmic ADP-ribosylome in these tumor types (Supplementary Fig. 8).
Discussion
In this study, we describe heterogeneous ADPR staining patterns in ovarian, lung, breast, prostate, and colorectal adenocarcinomas and different brain tumors by using a newly developed anti-ADP-ribose (ADPr) antibody. Loss of cyADPR signal was associated with advanced tumor stage, lymph node metastasis or shorter overall survival only in specific adenocarcinoma types.
PARP inhibitors are currently used in breast and ovarian cancer therapies [2, 50, 51] and considered in clinical studies for the treatment of other frequently diagnosed tumor types (e.g., brain, colon, lung, and prostate [7, 41,42,43,44,45,46,47]). PARPi target and inhibit some ARTs, among which PARP1/ARTD1 is the most abundant. Since PARP1/ARTD1 is mainly localized in the nucleus [6, 22], so far, the attention of the ADP-ribosylation studies was mostly on nuclear ADPR. In this study, we identified the presence of a homogeneous and strong nuclear staining in however a small number of breast cancer biopsies. A deeper analysis revealed in many nuclei the presence of foci in the presence of otherwise weak overall ADPR staining. The number of foci most likely reflects the activity of ARTs involved in nuclear ADPR, and therefore their quantification might be highly informative. Thus, an extended analysis with confocal microscopy and supported by specific softwares might reveal the prognostic relevance of ADPR nuclear foci in different cancer types in the future.
Despite of the importance of nuclear ADPR, we exclusively focused here on cyADPR. We provide evidence that cyADPR is reduced in advanced breast, ovarian, and colorectal cancer with higher stages and lymph node metastases. Therefore, strong cyADPR signal intensity correlated with a better prognosis in breast and high grade serous ovarian cancer (with a similar trend in lung adenocarcinoma). Although the prognostic relevance of cyADPR staining was lost in multivariate analysis, our data indicated that in some tumor types cyADPR levels decrease during tumor progression. The reason for this phenomenon might be connected to different factors. First, the alteration during tumor development of cell metabolism and NAD+ availability as ADPR reaction substrate, which was shown to exert a protective effect against oncogenesis [52, 53], and alternatively, the modulation during tumorigenesis of the presence/activity of enzymes responsible for ADPR. Expanded investigations about the NAD+ homeostasis and the enzyme expression levels will provide further insights for the importance of ADPR levels during cancer development.
A very recent study underlined a correlation between ADPR levels and clinical outcome in a small cohort (n = 34) of ovarian cancer patients [29]. Differently than what we observed, that study indicated a negative correlation between the PARP1/ARTD1-dependent ADPR levels and the clinical outcome of high grade serous ovarian cancer patients [29]. This difference might be linked to different factors besides the cohort size. First, the tools (antibody or binding reagent) used to assess ADPR levels were different and might prefer ADPr moieties linked to specific amino acid acceptor sites thus reporting only a partial overview of the cellular ADP-ribosylome. Second, the ADPR-detecting tools used in these two studies were employed with different methodologies: immunoblot or immunohistology. Unfortunately, since the ADPR binding reagents used in the immunoblot-based study were not validated for IHC, a direct comparison on patient histological sections was not possible. Third, cell lysate ADPR, analyzed in the other study, differs from cytoplasmic ADPR, since it includes also nuclear ADPR which is largely catalyzed by PARP1/ARTD1 and whose contribution to extranuclear ADPR remains to be investigated.
In this study, we also investigated the subcellular source of cyADPR. The cytoplasm consists of several compartments/organelles (e.g., mitochondria, endoplasmic reticulum, golgi, cytosol) that could contribute to cyADPR, each of them characterized by the presence of different enzymes regulating ADPR [23, 54, 55]. We observed that in breast, colon, lung, and ovarian carcinomas the major source of cyADPR was the mitochondrion. However, residual ADPR signal was observed not to colocalize with this specific organelle. Thus, in order to precisely understand cyADPR distribution in the cytoplasm, a future colocalization study should be considered. Defining the exact localization of all cyADPR will allow to further investigate the molecular mechanism regulating cyADPR levels and the various patient prognosis. Additional mechanistic data might come from mass spectrometry analysis of the ADP-ribosylome in biopsies displaying different cyADPR staining intensity.
In this comprehensive study, we considered different cancer types: breast, ovarian, colon, lung, prostate and brain. Although for brain and colon cancer patient survival data were not available, in breast and high grade serous ovarian cancer cyADPR correlated with patient OS, whereas no statistically significant association was found for prostate and lung cancer. The reason for this discrepancy might be the limited number of cases available for the analysis in some tumor (sub)types (i.e., lung squamous and adenocarcinoma). Thus, for potential follow-up studies larger patient cohorts are needed to assess the prognostic value of cyADPR in other tumor types.
The anti-ADPr antibody used for the IHC stainings presented in this study detected ADPR, but did not allow to distinguish between MAR or PAR. For future studies, it would be of great interest to understand the prognostic impact of these two ADPR forms. Whether the determination of ADPR patterns and forms in aggressive tumors would also provide a rational for a therapeutic intervention remains to be investigated, especially since the currently available PARPi target mainly the nuclear ADPR and no other ART family members.
In summary, we provide evidence that the pattern of cyADPR implies an important role for cyADPR in the behavior of specific tumor types. Our data might be important for the understanding of different responses to PARPi therapy. In this context, it is tempting to speculate whether, in addition to “BRCAness”, the assessment of nuclear and/or cytoplasmic ADPR levels represents a predictive diagnostic tool for cancer therapy with PARPi.
Data availability
All data is available in the main text or the supplementary materials. Raw data and materials are available upon request for non-commercial research purposes.
References
McCann KE, Hurvitz SA. Advances in the use of PARP inhibitor therapy for breast cancer. Drugs Context. 2018;7:212540.
Lyons TG, Robson ME. Resurrection of PARP inhibitors in breast cancer. J Natl Compr Canc Netw. 2018;16:1150–6.
LP, A. P. LYNPARZA® (olaparib) capsules, for oral use Initial U.S. Approval: 2014, https://www.accessdata.fda.gov/drugsatfda_docs/label/2018/206162s011lbl.pdf Accessed July 8, 2020.
AstraZeneca. Summary of product characteristics, 2014, https://www.ema.europa.eu/en/documents/product-information/lynparza-epar-product-information_en.pdf Accessed July 8, 2020.
Kamel D, Gray C, Walia JS, Kumar V. PARP inhibitor drugs in the treatment of breast, ovarian, prostate and pancreatic cancers: an update of clinical trials. Curr Drug Targets. 2018;19:21–37.
Caldecott KW. Protein ADP-ribosylation and the cellular response to DNA strand breaks. DNA repair. 2014;19:108–13.
Hussain M, Mateo J, Fizazi K, Saad F, Shore N, Sandhu S, et al. Survival with olaparib in metastatic castration-resistant prostate cancer. N. Engl J Med. 2020. https://doi.org/10.1056/NEJMoa2022485
Jiang Y, Dai H, Li Y, Yin J, Guo S, Lin SY, et al. PARP inhibitors synergize with gemcitabine by potentiating DNA damage in non-small-cell lung cancer. Int J Cancer. 2019;144:1092–103.
Zhu H, Wei M, Xu J, Hua J, Liang C, Meng Q, et al. PARP inhibitors in pancreatic cancer: molecular mechanisms and clinical applications. Mol Cancer. 2020;19:49.
de Koning L, Decaudin D, El Botty R, Nicolas A, Carita G, Schuller M, et al. PARP inhibition increases the response to chemotherapy in uveal melanoma. Cancers (Basel). 2019;11:751–67.
Pilie PG, Gay CM, Byers LA, O’Connor MJ, Yap TA. PARP inhibitors: extending benefit beyond brca-mutant cancers. Clin Cancer Res. 2019;25:3759–71.
Reilly NM, Novara L, Di Nicolantonio F, Bardelli A. Exploiting DNA repair defects in colorectal cancer. Mol Oncol. 2019;13:681–700.
Augustine T, Maitra R, Zhang J, Nayak J, Goel S. Sensitization of colorectal cancer to irinotecan therapy by PARP inhibitor rucaparib. Invest New Drugs. 2019;37:948–60.
Arena S, Corti G, Durinikova E, Montone M, Reilly NM, Russo M, et al. A subset of colorectal cancers with cross-sensitivity to olaparib and oxaliplatin. Clin Cancer Res. 2020;26:1372–84.
Mark C Markowski, M., Ph.D. Phase II study of olaparib in metastatic renal cell carcinoma patients harboring a BAP-1 or other DNA repair gene mutations (ORCHID), https://www.smartpatients.com/trials/NCT03786796 Accessed July 8, 2020, (2018).
Olson D, Bhalla S, Yang X, Martone B, Kuzel TM. Novel use of targeted therapy via PARP-inhibition in a rare form of papillary renal cell carcinoma: a case report and literature review. Clin Genitourin Cancer. 2016;14:e445–448.
Chen A. PARP inhibitors: its role in treatment of cancer. Chin J Cancer. 2011;30:463–71.
Byrum AK, Vindigni A, Mosammaparast N. Defining and modulating ‘BRCAness’. Trends Cell Biol. 2019;29:740–51.
Antolin AA, Ameratunga M, Banerji U, Clarke PA, Workman P, Al-Lazikani B. The kinase polypharmacology landscape of clinical PARP inhibitors. Sci Rep. 2020;10:2585.
Hottiger MO. Poly(ADP-ribose) polymerase inhibitor therapeutic effect: are we just scratching the surface? Expert Opin Ther Targets. 2015;19:1149–52.
Barkauskaite E, Jankevicius G, Ahel I. Structures and mechanisms of enzymes employed in the synthesis and degradation of PARP-dependent protein ADP-ribosylation. Mol Cell. 2015;58:935–46.
Luo X, Kraus WL. On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 2012;26:417–32.
Butepage M, Eckei L, Verheugd P, Luscher B. Intracellular mono-ADP-ribosylation in signaling and disease. Cells. 2015;4:569–95.
Yu M, Schreek S, Cerni C, Schamberger C, Lesniewicz K, Poreba E, et al. PARP-10, a novel Myc-interacting protein with poly(ADP-ribose) polymerase activity, inhibits transformation. Oncogene. 2005;24:1982–93.
Kaufmann M, Feijs KL, Luscher B. Function and regulation of the mono-ADP-ribosyltransferase ARTD10. Curr Top Microbiol Immunol. 2015;384:167–88.
Kistemaker HAV, Voorneveld J, Filippov DV. ADPr-peptide synthesis. Methods Mol Biol. 2018;1813:345–69.
Voorneveld J, Rack JGM, Ahel I, Overkleeft HS, van der Marel GA, Filippov DV. Synthetic alpha- and beta-Ser-ADP-ribosylated peptides reveal alpha-Ser-ADPr as the Native Epimer. Org Lett. 2018;20:4140–3.
Feijs KLH, Cooper CDO, Zaja R. The controversial roles of ADP-ribosyl hydrolases MACROD1, MACROD2 and TARG1 in carcinogenesis. Cancers (Basel). 2020;12:604–21.
Conrad LB, Lin KY, Nandu T, Gibson BA, Lea JS, Kraus WL. ADP-ribosylation levels and patterns correlate with gene expression and clinical outcomes in ovarian cancers. Mol Cancer Ther. 2020;19:282–91.
Moyle PM, Muir TW. Method for the synthesis of mono-ADP-ribose conjugated peptides. J Am Chem Soc. 2010;132:15878–80.
Xu Y, Zhou P, Cheng S, Lu Q, Nowak K, Hopp AK, et al. A bacterial effector reveals the V-ATPase-ATG16L1 axis that initiates xenophagy. Cell. 2019;178:552–66 e520.
Hopp, AK et al. Mitochondrial NAD+ controls nuclear ARTD1-induced ADP-ribosylation. Mol Cell.2021;340–54 https://doi.org/10.1016/j.molcel.2020.12.034
Probst-Hensch NM, Steiner JH, Schraml P, Varga Z, Zurrer-Hardi U, Storz M, et al. IGFBP2 and IGFBP3 protein expressions in human breast cancer: association with hormonal factors and obesity. Clin Cancer Res. 2010;16:1025–32.
Noske A, Henricksen LA, LaFleur B, Zimmermann AK, Tubbs A, Singh S, et al. Characterization of the 19q12 amplification including CCNE1 and URI in different epithelial ovarian cancer subtypes. Exp Mol Pathol. 2015;98:47–54.
Tischler V, Fritzsche FR, Wild PJ, Stephan C, Seifert HH, Riener MO, et al. Periostin is up-regulated in high grade and high stage prostate cancer. BMC Cancer. 2010;10:273.
Gachechiladze M, Skarda J, Skanderova D, Uberall I, Kolek V, Smickova P, et al. Prognostic value of tumor-infiltrating lymphocytes (TILs) and their association with PD-L1 expression and DNA repair protein RAD51 in patients with resected non-small cell lung carcinoma. Lung Cancer. 2020;147:30–38.
Mertz KD, Mager LF, Wasmer MH, Thiesler T, Koelzer VH, Ruzzante G, et al. The IL-33/ST2 pathway contributes to intestinal tumorigenesis in humans and mice. Oncoimmunology. 2016;5:e1062966.
Gramatzki D, Frei K, Cathomas G, Moch H, Weller M, Mertz KD. Interleukin-33 in human gliomas: Expression and prognostic significance. Oncol Lett. 2016;12:445–52.
Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4:844–7.
STHDA. Cox proportional-hazards model, http://www.sthda.com/english/wiki/cox-proportional-hazards-model. Accessed August 26th, 2020.
Mateo J, Porta N, Bianchini D, McGovern U, Elliott T, Jones R, et al. Olaparib in patients with metastatic castration-resistant prostate cancer with DNA repair gene aberrations (TOPARP-B): a multicentre, open-label, randomised, phase 2 trial. Lancet Oncol. 2020;21:162–74.
Ramakrishnan Geethakumari P, Schiewer MJ, Knudsen KE, Kelly WK. PARP Inhibitors in Prostate Cancer. Curr Treat Options Oncol. 2017;18:37.
Dai W, Fu Y, Deng Y, Zeng Z, Gu P, Liu H, et al. Regulation of wnt singaling pathway by poly (ADP-Ribose) glycohydrolase (PARG) silencing suppresses lung cancer in mice induced by benzo(a)pyrene Inhalation Exposure. Front Pharm. 2019;10:338.
Michels J, Adam J, Goubar A, Obrist F, Damotte D, Robin A, et al. Negative prognostic value of high levels of intracellular poly(ADP-ribose) in non-small cell lung cancer. Ann Oncol. 2015;26:2470–7.
Chornenkyy Y, Agnihotri S, Yu M, Buczkowicz P, Rakopoulos P, Golbourn B, et al. Poly-ADP-ribose polymerase as a therapeutic target in pediatric diffuse intrinsic pontine glioma and pediatric high-grade astrocytoma. Mol Cancer Ther. 2015;14:2560–8.
Zhang NN, Lin T, Xiao M, Li QS, Li X, Yang L, et al. Transcriptome sequencing analysis of monoADPribosylation in colorectal cancer cells. Oncol Rep. 2020;43:1413–28.
Palazzo L, Mikolcevic P, Mikoc A, Ahel I. ADP-ribosylation signalling and human disease. Open Biol. 2019;9:190041–59.
Dolle C, Rack JG, Ziegler M. NAD and ADP-ribose metabolism in mitochondria. FEBS J. 2013;280:3530–41.
Brunyanszki A, Szczesny B, Virag L, Szabo C. Mitochondrial poly(ADP-ribose) polymerase: the wizard of Oz at work. Free Radic Biol Med. 2016;100:257–70.
Franzese E, Centonze S, Diana A, Carlino F, Guerrera LP, Di Napoli M, et al. PARP inhibitors in ovarian cancer. Cancer Treat Rev. 2019;73:1–9.
O’Cearbhaill RE. Using PARP inhibitors in advanced ovarian cancer. Oncol (Williston Park). 2018;32:339–43.
Pramono AA, Rather GM, Herman H, Lestari K, Bertino JR. NAD- and NADPH-contributing enzymes as therapeutic targets in cancer: an overview. Biomolecules. 2020;10:358–76.
Poljsak B. NAD+ in cancer prevention and treatment: pros and cons. J Clin Exp Oncol. 2016. https://doi.org/10.4172/2324-9110.1000165
Hopp AK, Gruter P, Hottiger MO. Regulation of Glucose Metabolism by NAD(+) and ADP-Ribosylation. Cells. 2019;8:890–913.
O’Sullivan J, Tedim Ferreira M, Gagne JP, Sharma AK, Hendzel MJ, Masson JY, et al. Emerging roles of eraser enzymes in the dynamic control of protein ADP-ribosylation. Nat Commun. 2019;10:1182.
Acknowledgements
We thank Subham Mridha (Department of Quantitative Biomedicine, University of Zurich) for providing support with software coding and statistical analysis. We thank Susanne Dettwiler and Fabiola Prutek (Tissue Biobank, University Hospital Zurich) for TMA cutting and staining, as well as Christiane Mittmann and André Fitsche (In Situ Laboratory, Dept. of Pathology, University Hospital Zurich) for TMA staining optimization. We thank Elisabeth Rushing, University Hospital Zurich, for providing support with the analysis of brain tumors. We thank Tobias Suter (University of Zurich) for the helpful discussions, the operations leading to the generation of the ADPR antibody and for providing editorial assistance.
Funding
FA is supported by Cancer Research Center of the University of Zurich. ADP-ribosylation research in the laboratory of MOH is funded by the Kanton of Zurich and the Swiss National Science Foundation (grant 310030_176177). Open Access funding provided by Universität Zürich.
Author information
Authors and Affiliations
Contributions
Project conceptualization and administration by: MOH, FA (lead) and PS, HM (supporting). Investigation by: FA (lead), HM, PS, MOH (supporting). Methodology: FA (TMA staining optimization) and PS (TMA selection, cutting, and staining). Data curation and formal analysis by: FA (lead), PS (TMA data, supporting for TMA scoring). Visualization and validation by: FA (lead), HM and PS (independent control). Writing, review, and editing of the manuscript: MOH, FA (lead), PS (editing), HM (supporting).
Corresponding author
Ethics declarations
Conflict of interest
A patent application for the use of ADPR staining has be filed by the University of Zurich. The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Aimi, F., Moch, H., Schraml, P. et al. Cytoplasmic ADP-ribosylation levels correlate with markers of patient outcome in distinct human cancers. Mod Pathol 34, 1468–1477 (2021). https://doi.org/10.1038/s41379-021-00788-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-021-00788-9