Abstract
Rhabdomyosarcomas with TFCP2 fusions represent an emerging subtype of tumors, initially discovered by RNA-sequencing. We report herein the clinicopathological, transcriptional, and genomic features of a series of 14 cases. Cases were retrospectively and prospectively recruited and studied by immunohistochemistry (MYF4, MYOD1, S100, AE1/E3, ALK), fluorescence in situ hybridization with TFCP2 break-apart probe (n = 10/14), array-comparative genomic hybridization (Agilent), whole RNA-sequencing (Truseq Exome, Illumina), or anchored multiplex PCR-based targeted next-generation sequencing (Archer® FusionPlex® Sarcoma kit). Patient’s age ranged between 11 and 86 years, including 5 pediatric cases. Tumors were located in the bone (n = 12/14) and soft tissue (n = 2/14). Most bone tumors invaded surrounding soft tissue. Craniofacial bones were over-represented (n = 8/12). Median survival was 8 months and five patients are currently alive with a median follow-up of 20 months. Most tumors displayed a mixed spindle cell and epithelioid pattern with frequent vesicular nuclei. All tumors expressed keratins and showed a rhabdomyogenic phenotype (defined as expression of MYF4 and/or MYOD1). ALK was overexpressed in all but three cases without underlying ALK fusion on break-apart FISH (n = 5) nor next-generation sequencing (n = 14). ALK upregulation was frequently associated with an internal deletion at genomic level. TFCP2 was fused in 5′ either to EWSR1 (n = 6) or FUS (n = 8). EWSR1 was involved in both soft tissue cases. FISH with TFCP2 break-apart probe was positive in all tested cases (n = 8), including one case with unbalanced signal. On array-CGH, all tested tumors displayed complex genetic profiles with genomic indexes ranging from 13 to 107.55 and recurrent CDKN2A deletions. FET-TFCP2 rhabdomyosarcomas clustered together and distinctly from other rhabdomyosarcomas subgroups. Altogether, our data confirm and expand the spectrum of the new family of FET-TFCP2 rhabdomyosarcomas, which are associated with a predilection for the craniofacial bones, an aggressive course, and recurrent pathological features. Their association with ALK overexpression might represent a therapeutic vulnerability.
Similar content being viewed by others
Introduction
Rhabdomyosarcomas belong to a clinically and biologically heterogeneous family of tumors sharing re-expression of immature skeletal muscle differentiation factors, including MYF4/MYOGENIN and/or MYOD1 proteins. They are currently classified into four main subtypes: embryonal, alveolar, pleomorphic, and spindle cell/sclerosing [1]. Nonetheless, recent studies have deciphered the spindle cell/sclerosing family into molecularly distinct subtypes including NCOA2-rearranged infantile rhabdomyosarcomas [2], VGLL2-rearranged rhabdomyosarcomas [3], and spindle cell/sclerosing rhabdomyosarcomas with MYOD1 exon 1 mutations [4, 5].
More recently, a new subtype of rhabdomyosarcomas has been identified through an RNA-sequencing screening, defined by Transcription Factor Cellular Promoter 2 (TFCP2) rearrangements [6]. These tumors were initially reported as a “new variant of epithelioid rhabdomyosarcomas” and have a striking predilection for bones, especially for the craniofacial skeleton, which is a very uncommon feature for rhabdomyosarcomas [6]. Their morphological spectrum has been subsequently extended with the description of cases with spindle cell morphology in the small case series reported so far, with a total of ten cases now described in the literature [7,8,9,10]. The emerging evidence highlight their predilection for bones, female patients, a frequent spindle cell morphology, and common ALK overexpression but substantial clinical follow-up is available for only two cases.
We report herein the clinicopathological and molecular features of a series of 14 FET-TFCP2 rhabdomyosarcomas, highlighting their recurrent morphological and molecular features, paving the way to their diagnosis and recognition in routine practice. Our data confirm that they represent a distinct type of rhabdomyosarcoma.
Material and methods
Sample selection
Three index cases (TFCP2 1–3) were identified through a retrospective RNA-sequencing screening of unclassified sarcomas and were previously reported without detailed clinicopathological data [6]. Seven cases were identified prospectively: six cases were diagnosed on morphological grounds and one case (TFCP2-8) diagnosed on molecular grounds. Control cases were retrieved from the archives of the sarcoma pathology review network (RRePS and ResOs) and Institut Bergonie, including malignant peripheral nerve sheath tumors with rhabdomyosarcomatous differentiation (Triton tumor), and epithelioid/pleomorphic/embryonal and alveolar rhabdomyosarcomas. All sarcoma cases are recorded in the national sarcoma pathology RRePS and ResOs databases, approved by the National Committee for Protection of Personal Data (number 910390), in compliance with ethics principles of the chart of Helsinki. Cases 11–14 were retrieved from the files of AHGC and JVMGB, including two retrospective and two prospective cases.
Clinical review
Clinical follow-up was obtained from the medical records of patients provided by participating institutions or through corresponding clinicians. Survival was calculated from the date of diagnosis to the date of death or last date of follow-up. The clinical onset of disease was defined by the first symptom of cancer. Follow-up duration was calculated from the date of the first clinical consultation. Clinical management data are provided in Table 2. Survival analysis was performed with the Kaplan–Meier method using the dates of initial diagnosis and the dates of death from any cause.
Histopathological analysis
All material (biopsy and surgical specimen if surgery was performed) was centrally reviewed by specialized soft tissue and bone pathologists (FLL, JMC, AHGC, JVMGB).
Immunohistochemistry
The tissue slides were deparaffinized in xylene, hydrated in alcohol, and baked in a microwave (30 min in Tris buffer pH 9). Endogenous peroxidase was blocked. Staining was performed on the Benchmark ultra-automated stainer (Ventana) using diamino-benzidine as chromogen (Dako, Glostrup, Denmark). The following antibodies were used: AE1/E3 (clone PCK26, Ventana), KL1 (clone KL1, HISTOLS reagent), EMA (clone E29, Ventana), P63 (4A4, Ventana), S100 protein (clone poly Z311, Dako), SOX10 (EP268, BioSB), Desmin (clone DE-R11, Ventana), Myogenin (clone LO26, Leica), MYOD1 (clone EP212, Cell marque), ALK (clones D5F3, Cell Signaling Technology and clone 1A4, Novocastra), CD99/MIC2 (12E7, Dako), ETV4 (clone 16, Santa Cruz), CD30 (BerH2, Dako), CD4 (4B12, Novocastra), and CD7 (SP94, Ventana).
Fluorescence in situ hybridization (FISH)
Fluorescence in situ hybridization (FISH) analyses were performed with 4 μm sections of formalin-fixed paraffin-embedded (FFPE) tissue with the use of Histology FISH Accessory Kit (DAKO, reference: K5799) using commercial break-apart probes targeting EWSR1 (ZytoLight SPEC EWSR1 Dual Color Break Apart Probe, Zytovision, reference Z-2096–200), FUS (ZytoLight SPEC FUS Dual Color Break Apart Probe, Zytovision, reference Z-2130-50), ALK (ZytoLight SPEC ALK Dual Color Break Apart Probe, Zytovision, reference Z-2124-200), TFCP2 (TFCP2 Break Apart probe, Empire Genomics), and reference EG-TFCP2BA-20-GROR. Nuclei were scored for non-rearranged patterns (red and green fusion signals), rearranged, and unbalanced patterns (split of red and green signals or extra single red signals) using a Nikon Eclipse 80i fluorescent microscope with appropriate filters. The positive cutoff was ≥20%. Pictures were captured using a Hamamatsu C4742-95 CCD camera and analyzed with the Genikon software (Alphelys).
Array-comparative genomic hybridization
Genomic DNA was extracted from formalin-fixed paraffin-embedded tissue using QIAamp, DNA micro kit (Qiagen, Hilden, Germany). Genomic DNA and human reference DNA (Promega) were labeled with cyanin 5 and cyanin 3, respectively, using the Genomic DNA ULS Labeling Kit (Agilent Technologies, Santa Clara, California) and co-hybridized onto a Sureprint G3 Human CGH microarray 4 × 180 K (Agilent) following the manufacturer’s recommendations. Data were analyzed with Agilent Genomic Workbench software (v7.0, Agilent) or by Cytogenomics software (v2.9.2.4, Agilent). Large-size alterations were defined as quantitative anomalies of whole chromosomes or of segments of chromosomes involving many probes of the array, easily detectable without a specific query on a given genomic region. Small-size alterations were quantitative anomalies involving only few probes, not detected by the automatic algorithm but after manual analysis of the genomic regions.
Sanger sequencing of MYOD1
Fifty nanograms of FFPE DNA was amplified with forward (5′-AAGCGCAAGACCACCAAC-3′) and reverse primers (5′-GTTGCGCAGGATCTCCAC-3′) targeting exon 1 of MYOD1. The PCR products were analyzed by 2% agarose gel electrophoresis with an expected product size of 153 bp. The PCR products were sequenced using a Big Dye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) on a 3130XL Genetic Analyzer (Applied Biosystems).
Whole RNA-sequencing
RNA-sequencing was performed on formalin-fixed paraffin-embedded material in tencases (notably, cases TFCP2 1–3, the three previously reported cases studied by RNA-sequencing on frozen material [6], were re-sequenced with FFPE samples to ensure data comparability).
Total RNA was extracted from formalin-fixed paraffin-embedded tissue section using the Formapure RNA kit (Beckman Coulter, Brea, California) following the manufacturer’s recommendations. Quantity and quality of total RNA were evaluated using NanoDrop (Thermo Fisher Scientific) and Tape Station with Hs RNA Screen Tape (Agilent) using a cutoff of DV200 (defined as the percentage of RNA fragments above 200 nucleotides) above 30%. All samples passed quality criteria. Libraries were prepared with 100 ng of total RNA using TruSeq RNA Access Library Prep Kit (Illumina, San Diego, USA). Libraries were pooled by a group of 12 samples. Paired-end sequencing was performed using the NextSeq 500/550 High Output V2 kit (150 cycles) on Illumina NextSeq 500 platform (Illumina, San Diego, CA). To perform the clustering analysis, gene expression values were extracted using Kallisto v0.42.5 tool [11] with GENECODE release 23 genome annotation based on GRCh38 genome reference. Kallisto Transcripts Per Kilobase Million (TPM) expression values were transformed in log2(TPM + 2) and all samples were normalized together using the quantile method from the R limma package within R (version 3.1.1) environment. Clustering was performed with the R package Cluster v2.0.3 ConsensusClusterPlus v 1.46 [12] using 1000 permutations of 80% of both samples and genes. Agnes was used as the clustering algorithm with Pearson’s correlation distance and Ward’s clustering method.
Anchored multiplex PCR
Cases 11–14 were analyzed only by targeted RNA-sequencing. For the detection of translocations, total nucleic acid was isolated from five FFPE slides of 10 μm using the Tissue Preparation System (Siemens) after microdissection. After measurement of RNA quantity for FFPE material with Qubit fluorometric quantification system (Life Technologies), the target-enriched cDNA library was prepared with the Archer® FusionPlex® Sarcoma kit as per the manufacturer’s description. As described previously [13], reverse transcription of RNA was followed by end repair, adenylation, and universal half-functional adapter ligation of double-stranded cDNA fragments. This was followed by two rounds of low-cycle PCR with universal primers and gene-specific primers, covering 26 target genes that rendered the library fully functional for clonal amplification and sequencing using the Ion Proton™ or the Ion S5TM system. With the Archer analysis software (version 5.0) the produced libraries were analyzed for the presence of relevant fusions. Sequence quality was assessed by the following criteria: Quality control (QC) score of <30, a minimal total read number of 1.5 million with >7% unique fragments and >40% RNA reads.
Results
Clinical, radiological, and macroscopic findings
The age at diagnosis ranged from 11 to 86 years with a mean of 31 years (Table 1). There were five pediatric patients (defined as <18 years of age) and a slight female predominance was seen (eight female vs. six male patients).
Tumor size ranged from 20 to 150 mm with a median size of 60 mm. Tumors were most often located in the bones (n = 12/14) or soft tissue (n = 2/14). Bone tumors presented at locally advanced stage (Figs. 1, 3, 4 and Supplementary Fig. S1), invading surrounding tissues in all but one case (n = 11/12). Two cases involved two contiguous bones (n = 2/12). In one case, the tumor was bifocal within femoral bone (case TFCP2-7). Involved bones included the cranial bones (n = 3), bones of the orofacial complex (n = 6, mandible being the most commonly affected bone), vertebra, and long bone and sacrum (one case each). When performed, gross examination confirmed, in case of bone origin, destruction of cortical bone and extension in surrounding soft tissue (Supplementary Fig. S1). Regarding soft tissue tumors, locations included the peritoneum and inguinal area (the origin and precise level of extension of this case were not available, TFCP2-6).
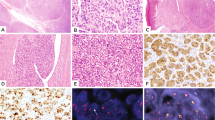
Typical case of FET-TFCP2 rhabdomyosarcoma. MR image demonstrating an expansive and destructive mass of the occipital bone in a 31yo male (a, TFCP2-9). The proliferation was arranged in solid sheets (b) and fascicles (c). Tumor cells displayed middle-sized nuclei with distinct nucleoli and mild anisocaryosis along with abundant eosinophilic cytoplasm (d). Cells stained focally for Myogenin (e), diffusely for MYOD1 (f), and diffusely for ALK (g). Cytokeratins AE1/3 and Desmin were heterogeneously and diffusely expressed, respectively (not shown)
Overall, all patients received chemotherapy with varied regimens, including two patients with neoadjuvant chemotherapy and four patients with chemotherapy as the sole therapeutic modality (Table 2). No consistent therapeutic response was achieved with conventional chemotherapy. Other treatment modalities included surgery (n = 7). Local control was achieved with surgery in six patients, including two R1 micro surgery (i.e., defined as the presence of microscopic tumor residue) and four R0 surgery but local relapse occurred in all patients (Table 2). Seven patients had metastatic disease, either at presentation (n = 3/14) or at follow-up (n = 4/14). Metastases involved lungs (n = 6/7), bones (n = 2/7), and lymph nodes (n = 1/7).
The mean survival was 8 months. The cohort included eight patients dead of disease and five patients alive, of whom two had evidence of clinical progression (Tables 1 and 2). The probability of survival was estimated to 46% at 12 months with Kaplan–Meier analysis (Supplementary Fig. S2). Among alive patients in this series (n = 5), all but one had undergone radical surgery and the mean follow-up was 6 months, with two patients currently in clinical progression. The sole survivor without surgery had unresectable tumor, which was progressive despite combined VDC-IE (vincristine–doxorubicin–cyclophosphamide–ifosfamide–etoposide) chemotherapy and radiation therapy, and was subsequently switched to targeted therapy (Alectinib), which halted progression (slight tumor regression from 53 × 27 to 32 × 29 mm) at 7 months of therapy (Table 2, TFCP2-8).
Microscopic findings
Tumors had been initially heterogeneously classified with diagnosis of spindle cell rhabdomyosarcoma (n = 3), epithelioid rhabdomyosarcoma (n = 2), embryonal and unclassified rhabdomyosarcomas not otherwise specified (NOS) (n = 2 and 2, respectively), malignant Triton tumor (n = 2), desmoplastic small round cell tumor (n = 1), unclassified sarcoma with a spindle cell phenotype (n = 1), and anaplastic lymphoma (n = 1) (Table 3).
Most tumors (n = 9/14) showed a hybrid spindle cell and epithelioid cytomorphology (Fig. 1b–d), whereas the other cases had essentially epithelioid (n = 3) (Fig. 2a, b), round cell (n = 1) (Fig. 2c, d), or spindle cell morphology (Fig. 3b, c).
Morphological variants seen in FET-TFCP2 rhabdomyosarcomas. Three FET-TFCP2 cases displayed pure epithelioid cytomorphology where tumor cells are arranged in solid sheets (a, case TFCP2-2) and harbored abundant globoid eosinophilic cytoplasm (b). One case of FET-TFCP2 rhabdomyosarcoma (TFCP2-8) displayed more abundant fibrous and myxoid stromal changes than seen in other cases (c) and tumor cells displayed small to medium rounded nuclei with fine chromatin without nucleoli (d). This tumor was associated with a FUS-TFCP2 fusion and displayed otherwise similar immunophenotypic and RNA expression features to other FET-TFCP2 tumors. FET-TFCP2 rhabdomyosarcomas may focally display fibrous stromal changes (e). Tumor nuclei were mostly monomorphic while a subset of cases harbored marked anisokaryosis with variations in size and shape of the nuclei (f)
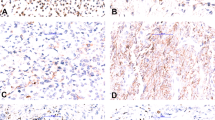
Case of FET-TFCP2 rhabdomyosarcoma with pure spindle cell moprhology. CT of an osteolytic lesion of the mandible with cortical destruction and soft-tissue extension (a, TFCP2-10). Tumor cells were arranged in intersecting fascicles with abundant eosinophilic cytoplasm (b, c). Tumor cells expressed heterogeneously desmin (d) and diffusely MYOD1 (e). Tumors cells displayed a split signal by FISH with the TFCP2 break-apart probe (f)
The lesional cells had abundant eosinophilic cytoplasm and oval nuclei dotted with distinct nucleoli in most cases (n = 9/14) (Fig. 4e). Nuclear hyperchromasia and pleomorphism (defined as variations in size and shape of the nuclei) were focally seen in four cases (Fig. 2f). Scattered rhabdomyoblasts were seen in two cases (n = 2/14) (Supplementary Fig. S1f). There was no significant stroma in half of the cases but focal fibrous changes were seen in six cases (Fig. 2e). A single case displayed a diffuse hyalinized stroma that encased the tumor cells, accounting for a trabecular growth pattern (Fig. 2c, d). Tumor necrosis was present in all but two tumors (Table 3).
Case of FET-TFCP2 rhabdomyosarcoma with small nuclear cytomorphology. CT of a multifocal osteolytic lesion of the right femur in the inter-trochanteric area and femoral neck (a, b, coronal reformatted and axial views, respectively) (case TFCP2-7). Core-needle biopsy evidence a mixed proliferation arranged in short fascicles (c) and solid sheets (d). Tumor cells harbor ovoid nuclei with fine chromatin without nucleoli, relatively monomorphic (e). The proliferation stained diffusely for ALK (f), AE1/E3 (g), and MYOD1 (h). A split signal is seen with the FISH assay using a break-apart probe spanning TFCP2 (i). Array-comparative genomic hybridization evidenced copy number alterations across the genome including homozygous deletion of CDKN2A and breakpoint within EWSR1 locus, in keeping with the presence of an underlying EWSR1-TFCP2 fusion (j)
Immunohistochemical findings
All but one case expressed desmin either diffusely (n = 8/14), heterogeneously (n = 3/14), or only focally (n = 2/14). All tumors expressed either MYOD1 or myogenin. MYOD1 was strongly expressed (i.e., >50%) in all but three cases (n = 11/14), whereas myogenin staining was only focally (<35%) positive in most cases (n = 9/14), negative in three cases (n = 3/14), and diffusely positive in two cases (Table 3 and Fig. 1). Notably, MYOD1 was diffusely positive in all tumors negative for myogenin. Keratins AE1/E3 were positive in all cases (n = 14), with focal staining in four cases (Figs. 1 and 4). ALK was overexpressed in all but three cases, with diffuse cytoplasmic overexpression (>50%) in all positive cases (Figs. 1, 4 and Supplementary Fig. S1). In one case, CD30 and CD4 were focally positive, accounting for its initial misdiagnosis as anaplastic lymphoma but no significant staining was seen in other tested cases (n = 8). S100 protein was focally positive in six cases and diffusely positive in one case, without concomitant expression of SOX10 in tested cases (n = 0/10).
Fluorescence in situ hybridization
EWSR1 break-apart FISH was positive in all but one EWSR1-TFCP2 tumors (n = 2/3), including one case with 3′ loss, consistent with unbalanced translocation (Supplementary Table S1). Break-apart FISH for FUS was positive in all tested cases (n = 5/5).
No rearrangement of ALK was seen nor by a FISH break part assay for ALK (n = 0/5) nor by RNA-sequencing (n = 0/14) (Supplementary Table S1).
FISH with TFCP2 break-apart probe was positive in all tested case (n = 9) (Figs. 3 and 4), including one case with an unbalanced split signal. To test the specificity of TFCP2 rearrangements in the setting of rhabdomyosarcomas, we tested 31 cases of rhabdomyosarcomas spanning the different subtypes by break-apart FISH with a break-apart probe for TFCP2 (Supplementary Table S2). All control cases were negative for rearrangement, including four sporadic malignant triton tumors, one neurofibromatosis type 1-associated malignant triton tumor, ten pleomorphic rhabdomyosarcomas, one case each of embryonal rhabdomyosarcoma and alveolar rhabdomyosarcoma, threespindle cell rhabdomyosarcomas, nine cases of epithelioid rhabdomyosarcomas, one case of carcinoma with rhabdomyosarcomatous component, and two cases of unclassified sarcomas with epithelioid phenotype (Supplementary Table S2).
Array-comparative genomic hybridization and MYOD1 mutational analysis
Mutations in the exon 1 of MYOD1 were absent at DNA level (n = 0/8) as well as at RNA level (n = 0/10) (Supplementary Table S1).
Genomic profiling was performed in ten cases, showing overall rearranged profiles with high genomic index ranging from 13.5 to 107.55. Two EWSR1-TFCP2 tumors (n = 2/4) and one FUS-TFCP2 tumor (n = 1/5) displayed genomic breakpoints within EWSR1 and FUS loci, respectively. Two tumors had a genomic breakpoint within TFCP2 locus. All cases displayed homozygous deletion of the tumor suppressor gene CDKN2A (n = 9/9) (Supplementary Table S3 and Fig. 4). Two cases displayed additional homozygous deletions spanning in the first case: PRKAR2A in 3p21.31, AUTS2 in 7q11.22, PTPN12 in 7q11.23-q21.11, PDE3A in 12p12.2, ST8SIA1 in 12p21.1, LARGE1 in 22q12.3, and in the second case MAP3K15, GPR63, and PHKA2 in Xp22.13-p22.12 and the locus 16p11.2-q12.1. Genomic amplifications were seen in one case spanning most notably MDM2 in 12q15, CCNE1 in 19q12-q13.11, and MAP3K1 in 5q11.2-12.1. Focusing on ALK locus, five cases displayed alterations (n = 5/9), including an internal hemizygous deletion in three cases, a hemizygous deletion, and a homozygous internal deletion in one case each.
RNA-sequencing findings
Whole RNA-sequencing was performed on FFPE material in ten cases and four cases were studied by Anchored multiplex PCR (Supplementary Table S1). Four cases of this series, including the three index cases of FET-TFCP2 rhabdomyosarcomas, were diagnosed based on RNA-sequencing findings without prior pathological suspicion of the diagnosis. TFCP2 was either fused to FUS (n = 8) or to EWSR1 (n = 6). Breakpoints involved constantly exon 2 of TFCP2, exons 5 or 6 of FUS and EWSR1. The transcriptomes of the seven cases sequenced on the same whole RNA-sequencing platform were compared with embryonal rhabdomyosarcomas (n = 6), alveolar rhabdomyosarcoma (n = 8), MYOD1-mutated rhabdomyosarcomas (n = 4), and VGLL2-fused rhabdomyosarcomas (n = 5). Unsupervised consensus clustering analyses highlighted that these seven FET-TFCP2 samples clustered apart from the other rhabdomyosarcomas (Supplementary Fig. S3). Expression analysis evidenced ALK RNA upregulation in all but two samples available for the analysis (n = 10). These two cases were notably negative for ALK in immunohistochemistry. ALK transcripts upregulation in FET-TFCP2 rhabdomyosarcomas was at least at the level of ALK-rearranged inflammatory myofibroblastic tumors or ALK-rearranged Spitz nevi (Supplementary Fig. S3). No fusion involving ALK was identified by targeted or whole transcriptome sequencing. At the transcriptional level, ALK upregulation was stronger in the subset of tumors associated with genomic deletion of ALK and was uneven along the transcript. The upregulation predominated on the sequences beyond exon 17, a finding which has been found in case of alternative transcription initiation [14] (Supplementary Fig. S4).
Fusions were characterized by anchored multiplex PCR in four cases, showing two cases each of FUS-TFCP2 and EWSR1-TFCP2 fusions, involving the same exons than those seen in the cohort explored by RNA-sequencing (Table 1).
Discussion
Rhabdomyosarcomas represent a clinically and biologically heterogeneous group of tumors that share by definition the re-expression of the immature myogenic markers Myogenin and MYOD1 [15]. However, rhabdomyosarcomatous differentiation can be seen in many tumor types, both epithelial with the example of carcinosarcomas [16] and mesenchymal malignancies, including malignant Triton tumors, biphenotypic sinonasal sarcomas [17], and mesenchymal chondrosarcomas [18]. The most well-defined categories of rhabdomyosarcomas are those associated with recurrent molecular alterations, which include alveolar rhabdomyosarcoma underlined by FOXO1 fusions [19], MYOD1-mutated spindle cell rhabdomyosarcomas [4, 5], and NCOA2- and VGLL2-fused spindle cell rhabdomyosarcomas [2, 3]. The identification of new recurrent molecular features may be helpful to break down the heterogeneous category of spindle cell rhabdomyosarcomas into homogeneous and clinically more accurate subtypes. Our initial description of FET-TFCP2 rhabdomyosarcomas [6] has hence been followed by the description of seven additional cases to date [6,7,8, 10, 20] (Table 4). Our series report 11 additional cases and provides a comprehensive clinical, pathological, genomic, and transcriptional characterization of this emerging entity.
Clinically, FET-TFCP2 rhabdomyosarcomas display a striking bone predilection with only two cases exclusively located in soft tissue in this series (Table 1), whereas no other soft tissue case is reported in the literature. Most bone cases destroyed the cortex and invaded surrounding soft tissue (Table 2). Most intriguingly, craniofacial bones are the commonest bones affected (n = 14/18). This is in keeping with available literature of intra-osseous rhabdomyosarcomas, as despite their extreme rarity, most published series are available in craniofacial bones, although without molecular characterization of the cases [21, 22]. Our data confirm that FET-TFCP2 tumors mostly affect young adult patients with a median age at diagnosis of 31 years with a slight female predominance (n = 13/21). Regarding outcomes, our series provides the best detailed clinical follow-up so far, with a median survival of 8 months. Among patients still alive, two have evidence of progressive disease (n = 2/5) despite the administration of multimodal therapy (Table 2) with available median follow-up of 20 months. Altogether, our series highlights the aggressive behavior of this entity.
Microscopically, in addition to our initial report, most FET-TFCP2 rhabdomyosarcomas actually display a hybrid spindle cell and epithelioid pattern. All tumors displayed high-grade features with vesicular nuclei, high mitotic activity, and tumor necrosis. A rhabdomyosarcomatous immunophenotype was always present, MYOD1 being more sensitive than Myogenin (Table 3). At transcriptional level, FET-TFCP2 tumors expressed high levels of skeletal muscle transcripts, especially sarcoglycan alpha, MYOD1, and MYOG [6]. All tumors consistently expressed keratins. Keratins are commonly expressed in rhabdomyosarcomas (staining was seen in 50% of alveolar rhabdomyosarcoma of our control cohort, Supplementary Table S4), although only focally, contrasting with the diffuse expression displayed by most FET-TFCP2 rhabdomyosarcomas (Table 3). Among the other published cases, one case with FUS-TFCP2 fusion is a “phenotypic outlier,” displaying a full myogenic profile (coexpression of caldesmon and smooth muscle actin (SMA)) and negative for keratins [10].
These tumors differ from the so-called “epithelioid rhabdomyosarcomas,” which display a pure solid architecture, a rare feature in FET-TFCP2 tumors with only three purely epithelioid cases in this series [23]. Moreover, reported cases of epithelioid rhabdomyosarcomas are located in deep soft tissue and scarcely express epithelial markers contrasting with the features of FET-TFCP2 rhabdomyosarcomas [23]. In the control cohort, all nine epithelioid rhabdomyosarcomas in soft tissue were negative for TFCP2 rearrangement by FISH break-apart assay, further supporting this pathological subtype is distinct from FET-TFCP2 rhabdomyosarcomas (Supplementary Table S2). We also report herein a new pattern in FET-TFCP2 rhabdomyosarcoma with one case associated with a round cell pattern, in which tumor cells were arranged in trabeculae and embedded in an abundant fibromyxoid stroma (Fig. 2c, d).
All cases were associated with TFCP2 rearrangements, fused in 5′ to FUS (n = 8/14) or EWSR1 (n = 6/14). TFCP2, also known as Late Simian Virus 40 factor (LSF), located in 12q13.12 encodes an evolutionarily conserved transcription factor that targets the thymidylate synthase gene, involved in the enzymatic regulation of DNA synthesis [24, 25]. TFCP2 contains an N-terminal DNA-binding domain and a C-terminal sterile alpha motif domain involved in protein dimerization [26] TFCP2 has been formerly linked to cancer with evidence of oncogenic deregulation in hepatocellular carcinoma [27] mediated by C-MET signaling [28]. Breakpoints within TFCP2 occur before the second exon, thereby preserving the DNA-binding domain of the protein. This fusion provides a further illustration of alternative involvement of EWSR1 or FUS in a single tumor entity, as these two genes are structurally and functionally related [29].
Our data show that FET-TFCP2 rhabdomyosarcomas are associated with overall complex genetic profiles. This is in agreement with the features reported in one case characterized by genomic profiling (Affymetrix platform), which showed a tetraploid profile with numerous alterations, chromotripsis of 1p and 3p, and homozygous deletion of CDKN2A [20]. CDKN2A inactivation was indeed seen in all tested cases of our series (Supplementary Table S3). Concomitantly, three cases had genomic features consistent with unbalanced translocation.
Despite their complex genomic rearrangements, the FET-TFCP2 rhabdomyosarcomas are transcriptionally homogeneous, as they clustered altogether. Moreover, adding more FET-TFCP2 samples to the initial clustering analyses strengthen their clustering away from the other rhabdomyosarcoma subtypes, as shown by the clear consensus clustering (Supplementary Fig. S3).
In this series, all but three FET-TFCP2 rhabdomyosarcomas overexpressed ALK both at transcriptional and protein levels, without underlying ALK fusion. According to RNA-sequencing and comparative genomic hybridization (CGH) data, this upregulation does not involve translocation nor amplification but correlated with ALK genomic deletion, presumably involving alternative transcription initiation [14] (Supplementary Fig. S4). No copy number alteration of ALK was detected in both FET-TFCP2 tumors negative for ALK by immunohistochemistry studied by array-CGH. Interestingly, ALK deletion has been previously reported in one case of FUS-TFCP2 sarcoma: the sequencing data showed that the upregulation did not involve exons 1–16, therefore in keeping with our finding [20]. Intriguingly, the expression of ALK has been shown with variable frequencies in other types of rhabdomyosarcomas, especially the alveolar subtype in up to 69% of cases [30, 31], but its underlying biological mechanism remains elusive. We confirmed this finding in our control cohort, as ALK staining was seen in 88.9% and 28.6% of alveolar and embryonal rhabdomyosarcomas, respectively (Supplementary Table S4). Nonetheless, ALK expression is rare in spindle cell rhabdomyosarcomas, present only in 23% of our control cases (Supplementary Table S4). Despite ALK overexpression, targeted therapy against ALK has not shown in vivo effects nor clinical benefits in alveolar rhabdomyosarcomas patients [32, 33], although the actual levels of expression of ALK were not reported in the studies. It is notable that at transcriptional level, the intensity of ALK upregulation is at least as high as in ALK-rearranged tumor types, such as ALK-fused inflammatory myofibroblastic tumors. Interestingly, one prospective patient of this series benefited from an ALK inhibitor after clinical progression under conventional chemotherapy, which stabilized tumor size with available follow-up of 15 months under therapy (Table 2). Therefore, the potential benefits of ALK inhibition in FET-TFCP2 rhabdomyosarcoma need to be further addressed, either as single therapy or in combination.
FET-TFCP2 rhabdomyosarcomas should not be confused with sarcomas with EWSR1-PATZ1 fusions, as these tumors may develop in the bones and display a rhabdomyogenic phenotype [34]. The latter tumors have a hybrid spindle cell to round cell phenotype but lack the epithelioid and high-grade nuclear features of most FET-TFCP2 rhabdomyosarcomas. FET-TFCP2 tumors, especially those that have an exclusive spindle cell morphology, may be confused with inflammatory myofibroblastic tumors. Both tumor types commonly express keratins and ALK. Nonetheless, ALK overexpression in inflammatory myofibroblastic tumors is related to underlying ALK fusion with a variety of partner genes, in contrast with FET-TFCP2 rhabdomyosarcomas. Intra-osseous rhabdomyosarcomas with MEIS1-NCOA2 fusions represent another emerging entity with only two cases reported for now [8]. This variant displays pure spindle cell morphology and does not express keratins nor ALK [8]. The rhabdomyogenic phenotype and focal staining for S100 protein seen in a subset of cases may also raise suspicion for a malignant Triton tumor, although none of TFCP2 patients had an underlying neurofibromatosis condition and the SOX10 marker was negative in all tested cases. Furthermore, we did not find TFCP2 fusions in control cases of malignant triton tumors (Supplementary Table S2).
In short, we report herein the largest and most comprehensive study of FET-TFCP2 rhabdomyosarcomas. This new entity is associated with hybrid epithelioid and spindle cell features, a predilection for craniofacial bones, and expresses consistently keratins and ALK. Tumors harboring these features should therefore benefit from molecular investigations seeking for a FET-TFCP2 fusion gene. Altogether, the clinical, morphological, and molecular data strongly suggest that these tumors represent a distinct rhabdomyosarcoma subtype.
References
Barr FG, Montgomery EA, Nascimento AF, Parham DM. Skeletal-muscle tumours. In: Fletcher CDM, Bridge JA, Hogendoorn PCW, Mertens F, editors. WHO classification of tumours of soft tissue and bone. 4th ed. Lyon: IARC Press; 2013. p.124–135.
Mosquera JM, Sboner A, Zhang L, Kitabayashi N, Chen CL, Sung YS, et al. Recurrent NCOA2 gene rearrangements in congenital/infantile spindle cell rhabdomyosarcoma. Genes Chromosomes Cancer. 2013;52:538–50.
Alaggio R, Zhang L, Sung YS, Huang SC, Chen CL, Bisogno G, et al. A molecular study of pediatric spindle and sclerosing rhabdomyosarcoma: identification of novel and recurrent VGLL2-related fusions in infantile cases. Am J Surg Pathol. 2016;40:224–35.
Kohsaka S, Shukla N, Ameur N, Ito T, Ng CK, Wang L, et al. A recurrent neomorphic mutation in MYOD1 defines a clinically aggressive subset of embryonal rhabdomyosarcoma associated with PI3K-AKT pathway mutations. Nat Genet. 2014;46:595–600.
Szuhai K, de Jong D, Leung WY, Fletcher CD, Hogendoorn PC. Transactivating mutation of the MYOD1 gene is a frequent event in adult spindle cell rhabdomyosarcoma. J Pathol. 2014;232:300–7.
Watson S, Perrin V, Guillemot D, Reynaud S, Coindre JM, Karanian M, et al. Transcriptomic definition of molecular subgroups of small round cell sarcomas. J Pathol. 2018;245:29–40.
Dashti NK, Wehrs RN, Thomas BC, Nair A, Davila J, Buckner JC, et al. Spindle cell rhabdomyosarcoma of bone with FUS-TFCP2 fusion: confirmation of a very recently described rhabdomyosarcoma subtype. Histopathology. 2018;73:514–20.
Agaram NP, Zhang L, Sung YS, Cavalcanti MS, Torrence D, Wexler L, et al. Expanding the spectrum of intraosseous rhabdomyosarcoma: correlation between 2 distinct gene fusions and phenotype. Am J Surg Pathol. 2019;43:695–702.
Tagami Y, Sugita S, Kubo T, Iesato N, Emori M, Takada K, et al. Spindle cell rhabdomyosarcoma in a lumbar vertebra with FUS-TFCP2 fusion. Pathol Res Pract. 2019; 205:152399 https://doi.org/10.1016/j.prp.2019.03.027.
Zhu G, Benayed R, Ho C, Mullaney K, Sukhadia P, Rios K, et al. Diagnosis of known sarcoma fusions and novel fusion partners by targeted RNA sequencing with identification of a recurrent ACTB-FOSB fusion in pseudomyogenic hemangioendothelioma. Mod Pathol. 2018;32:609–20.
Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–7.
Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–3.
Lam SW, Cleton-Jansen AM, Cleven AHG, Ruano D, van Wezel T, Szuhai K, et al. Molecular analysis of gene fusions in bone and soft tissue tumors by anchored multiplex PCR-based targeted next-generation sequencing. J Mol Diagn. 2018;20:653–63.
Wiesner T, Lee W, Obenauf AC, Ran L, Murali R, Zhang QG, et al. Alternative transcription initiation leads to expression of a novel ALK isoform in cancer. Nature. 2015;526:453–7.
Agaram NP. Update on myogenic sarcomas. Surg Pathol Clin. 2019;12:51–62.
McCluggage WG. A practical approach to the diagnosis of mixed epithelial and mesenchymal tumours of the uterus. Mod Pathol. 2016;29(Suppl 1):S78–91.
Le Loarer F, Laffont S, Lesluyes T, Tirode F, Antonescu C, Baglin AC, et al. Clinicopathologic and molecular features of a series of 41 biphenotypic sinonasal sarcomas expanding their molecular spectrum. Am J Surg Pathol. 2019;43:747–54 https://doi.org/10.1097/PAS.0000000000001238 (Epub).
Folpe AL, Graham RP, Martinez A, Schembri-Wismayer D, Boland J, Fritchie KJ. Mesenchymal chondrosarcomas showing immunohistochemical evidence of rhabdomyoblastic differentiation: a potential diagnostic pitfall. Hum Pathol. 2018;77:28–34.
Galili N, Davis RJ, Fredericks WJ, Mukhopadhyay S, Rauscher FJ, Emanuel BS, et al. Fusion of a fork head domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma. Nat Genet. 1993;5:230–5.
Wong DD, van Vliet C, Gaman A, Giardina T, Amanuel B. Rhabdomyosarcoma with FUS re-arrangement: additional case in support of a novel subtype. Pathology. 2019;51:116–20.
Iatrou I, Theologie-Lygidakis N, Schoinohoriti O, Tzermpos F, Vessala AM. Rhabdomyosarcoma of the maxillofacial region in children and adolescents: report of 9 cases and literature review. J Craniomaxillofac Surg. 2017;45:831–8.
Lucas DR, Ryan JR, Zalupski MM, Gross ML, Ravindranath Y, Ortman B. Primary embryonal rhabdomyosarcoma of long bone. Case report and review of the literature. Am J Surg Pathol. 1996;20:239–44.
Jo VY, Mariño-Enríquez A, Fletcher CD. Epithelioid rhabdomyosarcoma: clinicopathologic analysis of 16 cases of a morphologically distinct variant of rhabdomyosarcoma. Am J Surg Pathol. 2011;35:1523–30.
Veljkovic J, Hansen U. Lineage-specific and ubiquitous biological roles of the mammalian transcription factor LSF. Gene. 2004;343:23–40.
Swendeman SL, Spielholz C, Jenkins NA, Gilbert DJ, Copeland NG, Sheffery M. Characterization of the genomic structure, chromosomal location, promoter, and development expression of the alpha-globin transcription factor CP2. J Biol Chem. 1994;269:11663–71.
Kotarba G, Krzywinska E, Grabowska AI, Taracha A, Wilanowski T. TFCP2/TFCP2L1/UBP1 transcription factors in cancer. Cancer Lett 2018;28:72–79.
Yoo BK, Emdad L, Gredler R, Fuller C, Dumur Cl, Jones KH, et al. Transcription factor Late SV40 Factor (LSF) functions as an oncogene in hepatocellular carcinoma. Proc Natl Acad Sci USA. 2010;107:8357–62.
Yoo BK, Gredler R, Chen D, Santhekadur PK, Fisher PB, Sarkar D, et al. c-Met activation through a novel pathway involving osteopontin mediates oncogenesis by the transcription factor LSF. J Hepatol. 2011;55:1317–24.
Luo Y, Blechingberg J, Fernandes AM, Li S, Fryland T, Borglum AD, et al. EWS and FUS bind a subset of transcribed genes encoding proteins enriched in RNA regulatory functions. BMC Genomics. 2015;16:929.
Yoshida A, Shibata T, Wakai S, Ushiku T, Tsuta K, Fukayama M, et al. Anaplastic lymphoma kinase status in rhabdomyosarcomas. Mod Pathol. 2013;26:772–81.
Pillay K, Govender D, Chetty R. ALK protein expression in rhabdomyosarcomas. Histopathology. 2002;41:461–7.
Wierdl M, Tsurkan L, Chi L, Hatfield MJ, Tollemar V, Bradley C, et al. Targeting ALK in pediatric RMS does not induce antitumor activity in vivo. Cancer Chemother Pharm. 2018;82:251–63.
Schoffski P, Wozniak A, Leahy MG, Aamdal S, Rutkowski P, Bauer S, et al. The tyrosine kinase inhibitor crizotinib does not have clinically meaningful activity in heavily pre-treated patients with advanced alveolar rhabdomyosarcoma with FOXO rearrangement: European Organisation for Research and Treatment of Cancer phase 2 trial 90101 ‘CREATE’. Eur J Cancer. 2018;94:156–67.
Chougule A, Taylor MS, Nardi V, Chebib I, Cote GM, Choy E, et al. Spindle and round cell sarcoma with EWSR1-PATZ1 gene fusion: a sarcoma with polyphenotypic differentiation. Am J Surg Pathol 2019;43:220–8.
Acknowledgements
We are grateful to pathologists and clinicians for sharing cases and follow-up informations: Dr A Al Bouzidi (Rabat, Marocco), Dr E Angot (CHU Rouen), Dr Foucard (Paris), Professor C Berger (HEGP, Paris), Dr N Bennani Guebessi (Casablanca, Marocco), Dr C Bazille (CHU Caen), Professor F Gouin (CHU Nantes), Dr E Bompas (ICO, St Herblain), Dr Haffadi (Casablanca), Dr C Raban (CHU Poitiers), Dr V Laurence (Institut Curie, Paris), Dr A Moreau (CHU Nantes, France), Dr S Watson (Institut Curie, Paris, France), Professor D Baumhoer (Basel, Switzerland), Dr M Chan (Paramaribo, Suriname), and Dr U Flucke (Nijmegen, The Netherlands). Dr A de La Fouchardière (Centre Leon Berard, Lyon, France) graciously provided the ALK-fusion positive Spitz nevi for the transcriptional analysis. We thank L Mayeur, V Velasco, and V Fouqué for their assistance. INSERM U1218 is supported by a charity funding “Le Fil d’Oriane”.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Le Loarer, F., Cleven, A.H.G., Bouvier, C. et al. A subset of epithelioid and spindle cell rhabdomyosarcomas is associated with TFCP2 fusions and common ALK upregulation. Mod Pathol 33, 404–419 (2020). https://doi.org/10.1038/s41379-019-0323-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-019-0323-8
This article is cited by
-
Molecularly defined sinonasal malignancies: an overview with focus on the current WHO classification and recently described provisional entities
Virchows Archiv (2024)
-
TFCP2 as a therapeutic nexus: unveiling molecular signatures in cancer
Cancer and Metastasis Reviews (2024)
-
Molecular pathology in diagnosis and prognostication of head and neck tumors
Virchows Archiv (2024)
-
Multi-omic and functional analysis for classification and treatment of sarcomas with FUS-TFCP2 or EWSR1-TFCP2 fusions
Nature Communications (2024)
-
Epithelioid and spindle rhabdomyosarcoma with TFCP2 rearrangement in abdominal wall: a distinctive entity with poor prognosis
Diagnostic Pathology (2023)