Abstract
UTX/KDM6A, a histone H3K27 demethylase and a key component of the COMPASS complex, is frequently lost or mutated in cancer; however, its tumor suppressor function remains largely uncharacterized in multiple myeloma (MM). Here, we show that the conditional deletion of the X-linked Utx in germinal center (GC) derived cells collaborates with the activating BrafV600E mutation and promotes induction of lethal GC/post-GC B cell malignancies with MM-like plasma cell neoplasms being the most frequent. Mice that developed MM-like neoplasms showed expansion of clonal plasma cells in the bone marrow and extramedullary organs, serum M proteins, and anemia. Add-back of either wild-type UTX or a series of mutants revealed that cIDR domain, that forms phase-separated liquid condensates, is largely responsible for the catalytic activity-independent tumor suppressor function of UTX in MM cells. Utx loss in concert with BrafV600E only slightly induced MM-like profiles of transcriptome, chromatin accessibility, and H3K27 acetylation, however, it allowed plasma cells to gradually undergo full transformation through activation of transcriptional networks specific to MM that induce high levels of Myc expression. Our results reveal a tumor suppressor function of UTX in MM and implicate its insufficiency in the transcriptional reprogramming of plasma cells in the pathogenesis of MM.
Similar content being viewed by others
Introduction
Deregulation of histone modifications has been attributed to an imbalance between writers that install the histone methylation marks and erasers that remove these marks [1]. Specifically, alteration of the methylation of lysine 27 on histone H3 (H3K27) has an incriminating role in hematological malignancies [2, 3]. The catalytic components of polycomb repressive complex 2 (PRC2), enhancer of zeste homolog 2 (EZH2) and its homolog EZH1, deposit the repressive H3K27me3 mark at both promoters and enhancers [4]. The histone demethylase, ubiquitously transcribed tetratricopeptide repeat gene on the X chromosome (UTX), also known as KDM6A, specifically removes di- and tri-methyl groups from H3K27 thus antagonizing PRC2-mediated transcriptional repression [5]. UTX was identified as a critical regulator of reprogramming in induced pluripotent stem cells (iPSCs) through its catalytic activity by partnering with Oct4, Sox2, and KIF4 reprogramming factors [6]. In addition to its H3K27 demethylase activity, UTX possesses catalytic-independent functions. It contributes to embryonic viability, differentiation, and certain aspects of development in mouse embryonic stem cells (ESCs) independently of its demethylase activity [7,8,9]. Importantly, UTX is an integral component of the MLL3 and MLL4 COMPASS (complex of proteins associated with Set1) complexes, which monomethylate H3K4 at enhancers [10]. UTX mediates the conversion of enhancers from an inactive to an active state by direct recruitment and coupling of p300/CBP histone acetyltransferases (HAT) and MLL4 [11].
Recent studies revealed that UTX is frequently mutated in different kinds of cancer, including hematological malignancies; however, the pathological role of UTX mutations in tumorigenesis is not yet fully understood [12]. UTX is often inactivated, exclusively in males, in T-cell acute lymphoblastic leukemia (T-ALL), where it functions as a tumor suppressor [13, 14]. UTX also suppresses the development of myeloid leukemia, in which it functions independently of its demethylase activity, through remodeling of enhancers and chromatin accessibility of oncogenes and tumor suppressor genes [15]. In multiple myeloma (MM), UTX inactivating mutations/deletions were detected in 1.5–4% of patients [16,17,18] and more frequently in established cell lines [19], suggesting a tumor suppressor role for UTX in MM and its implication in disease progression. Importantly, UTX inactivating lesions were associated with adverse overall survival [17]. UTX-null MM cell lines were more sensitive to EZH2 inhibitors compared to UTX-wild type (WT) MM [19].
To investigate the role of UTX insufficiency in the pathogenesis of MM, we generated a novel mouse model that had concurrent Utx loss and the activating BrafV600E mutation. The activating mutations in the RAS/RAF/MEK/ERK/MAPK pathway are identified in up to 50% of newly diagnosed MM patients [16], and the serine-threonine kinase BRAF is mutated in 8-12% of MM patients at diagnosis, with V600E as the most common BRAF mutation [16, 20]. In this study, we demonstrated the tumor suppressor function of Utx in mature B cell malignancies including MM.
Materials and methods
Mice
All animal procedures were conducted in accordance with the Chiba University guidelines for the use of laboratory animals and approved by the Review Board for Animal Experiments of Chiba University (approval ID: 30-56) and Tokyo University (approval ID: PS18-02). Mice with conditional expression of BrafV600E (LSL-Braf V600E) from its endogenous locus were previously described [21]. Utx conditional knockout mice were recently reported [22]. Utxfl;BrafV600E mice were backcrossed at least 6 times onto a C57BL/6 (CD45.2) background and were crossed to Cγ1-Cre mice [23]. Cγ1-Cre-negative mice were used as controls. We immunized six- to eight-week-old mutant Utx/Braf, Cγ1Cre+ and control mice using intraperitoneal injection of 50 µg NP-CGG (Biosearch Technologies) emulsified in complete Freund’s adjuvant (Sigma) and then 4–6 weeks later we gave them a second dose of NP-CGG in incomplete Freund’s adjuvant (Sigma). NOD.Cg-PrkdcscidIl2rgtm1Sug/Jic (NOG) and NOD.Cg-PrkdcscidIl2rgtm1Sug Tg (CMV-IL6)/Jic (NOG-hIL-6) [24] mice were purchased from Central Institute for Experimental Animals (CIEA, Kawasaki, Japan). C57BL/6 mice congenic for the Ly5 locus (CD45.1) mice were purchased from Sankyo-Lab service, Tokyo, Japan.
Results
Utx loss cooperates with Braf V600E to induce B-cell neoplasms
To conditionally delete Utx in GC B cells, we crossed Utxfl mice, in which exons 11 and 12 were floxed [22], with Cγ1-Cre mice, which express Cre recombinase in GC B cells, class switched memory B cells, and plasma cells in response to immunization [23]. The Utx allele that lacks exons 11 and 12 encoding a part of the TPR domain generates a frame shift mutant that fails to code for the C-terminal domains including the JmjC catalytic domain [22]. We crossed Utxfl mice with LSL-BrafV600E mice, in which the BrafV600E allele is expressed from the endogenous Braf locus after Cre-mediated deletion of a lox-stop-lox (LSL) cassette [21]. We confirmed the efficient conditional deletion of Utx and LSL cassette using genomic polymerase chain reaction (PCR) of DNA extracted from spleen plasma cells (Supplementary Fig. 1A). Utx mRNA levels were significantly reduced in Utx-deficient plasma cells compared to control, possibly because of the activation of the nonsense-mediated mRNA decay pathway (Supplementary Fig. 1B). We hereafter refer to Cγ1-Cre;Utxfl/fl;BrafV600E/+, Cγ1-Cre;Utxfl/+;BrafV600E/+, and Cγ1-Cre;Utxfl/Y;BrafV600E/+ mice as UtxΔ/ΔBrafV600E, UtxΔ/+BrafV600E, and UtxΔ/YBrafV600E, respectively (Fig. 1A).
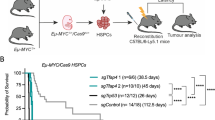
A Schematic diagram of experimental strategy. B Left panel: Kaplan–Meier survival analysis of Cre negative (n = 20), UtxΔ/y (n = 37), UtxΔ/+ (n = 19), UtxΔ/Δ (n = 37), BrafV600E (n = 16), UtxΔ/yBrafV600E (n = 33), UtxΔ/+BrafV600E (n = 18), and UtxΔ/ΔBrafV600E (n = 21) mice. Statistical significance of survival difference was determined by the log-rank test. **P < 0.01; and ***P < 0.001. Right panel: incidence of plasma cell neoplasms, B-cell lymphoma, and lymphoproliferative disease (LPD) in mice analyzed (UtxΔ/yBrafV600E, n = 20; UtxΔ/+BrafV600E, n = 16; UtxΔ/Δ BrafV600E, n = 17). C Spleen weights normalized to body weight (left) and hemoglobin levels in PB (right) from mice that developed plasma cell neoplasms (PCNs, n = 21 and 23, respectively), B-cell lymphoma (BCL, n = 11 each) and lymphoproliferative disease (LPD, n = 12 each), and Cre-negative control mice (Cre-, n = 16 and 26, respectively). Bars indicate mean. **P < 0.01; ***P < 0.001 by student t-test.
During a long-term observation period, UtxΔ/+ females did not develop any lethal diseases, while UtxΔ/Δ female and UtxΔ/Y males developed lethal B cell malignancies, including plasma cell neoplasms, B-cell lymphoma, and lymphoproliferative disease (LPD) at low frequencies (Fig. 1B left panel and Supplementary Table 1). BrafV600E mice also developed LPD and plasma cell neoplasms at low frequencies after a long latency (Fig. 1B left panel and Supplementary Table 1). Importantly, concurrent Utx loss and BrafV600E expression significantly shortened the survival of mice compared to Cre negative control, single Utx loss, and single BrafV600E expression (Fig. 1B left panel). UtxΔ/ΔBrafV600E females succumbed to disease earlier than UtxΔ/YBrafV600E males and UtxΔ/+BrafV600E females. Notably, UtxΔ/ΔBrafV600E, UtxΔ/YBrafV600E, and UtxΔ/+BrafV600E mice displayed heterogeneous phenotypes that included plasma cell neoplasms, B cell lymphoma, and LPD, with plasma cell neoplasms having the highest frequencies in UtxΔ/ΔBrafV600E and UtxΔ/YBrafV600E mice (Fig. 1B right panel). Among the compound mice, UtxΔ/YBrafV600E male mice developed plasma cell neoplasms at a higher frequency than compound heterozygous and homozygous Utx females (Fig. 1B right panel). Sick mice showed splenomegaly and advanced anemia irrespective of the disease type (Fig. 1C).
Development of B-cell lymphoma in Utx insufficient Braf V600E mice
A significant number of UtxΔ/YBrafV600E, UtxΔ/+BrafV600E, and UtxΔ/ΔBrafV600E mice developed B cell lymphoma with marked lymphadenopathy, splenomegaly, and hepatic involvement (25%, 6%, and 29%, respectively) (Fig. 1B right panel and Supplementary Table 2). Pathological examination of the spleen from UtxΔ/ΔBrafV600E (BU-159) mouse revealed expansion of B220+BCL6+ GC B-lymphocytes, giving a picture resembling follicular lymphoma (FL) (Fig. 2A left panel). Flow cytometric analysis of a mesenteric LN from the same mouse confirmed clonal expansion of GC B cells (B220+CD95+GL7+) (Fig. 2A right panel). Immunohistochemical analyses of the spleen and lymph nodes (LNs) from BU-322 and BU-434 mice revealed loss of the normal architecture with follicular and diffuse expansion of neoplastic B220+Pax5+ B-lymphocytes, showing FL- and diffuse large B cell lymphoma (DLBCL)-like appearance, respectively (Fig. 2B left panel). Flow cytometric analysis confirmed the clonal B cell expansion in the spleen and enlarged LNs (Fig. 2B right panel). Detailed flow cytometric analysis of B-cell subsets in the spleens of lymphoma mice showed a decrease in the percentage of follicular B cells (FO; B220+CD23+CD21low) and an increase in transitional B cells (TR; B220+CD23−CD21−) or GC B cells (Supplementary Table 2).
A Left panel: representative spleen sections harvested from a 40-week-old UtxΔ/ΔBrafV600E mouse (BU-159) stained with hematoxylin and eosin or antibodies recognizing the indicated marker antigens. Right panel: representative plots of flow cytometric analysis of GC B cells in a mesenteric lymph node of the same mouse. Cells were first gated on live (PI−) CD45.2+ B220+ B-lymphocytes to determine the percentage of GC B cells (CD95+GL7+) and then we evaluated the clonality using IgG1, κ, and λ antibodies. B Left panel: representative spleen (upper) and mesenteric lymph node (LN) (lower) sections harvested from 45- and 40-week-old UtxΔ/ΔBrafV600E mice (BU-322 and BU-434), respectively, stained with hematoxylin and eosin or antibodies recognizing the indicated marker antigens. Right panel: representative plots of flow cytometric analysis of GC B cells in the spleen and mediastinal or mesenteric lymph nodes of the same mice. We evaluated the clonality using IgA, κ, and λ antibodies.
While UTX mutations are rare in GC B-cell lymphomas [25], a high incidence of inactivating mutations in the components of COMPASS and its partner HATs, including KMT2D, CREBBP, and EP300, has been reported [26, 27]. Correspondingly, the deletion of Kmt2d, Crebbp, and Ep300 in GC B cells has been found to promote lymphomagenesis in mice [28,29,30]. UTX deletion in Eμ-Myc mice not only accelerates lymphomagenesis, but also promotes tumor progression [31]. In addition, lymphomas with low UTX expression are associated with poor patient survival [31]. These findings suggest that Utx loss in GC B cells deregulates COMPASS/HAT, thereby promoting lymphomagenesis in mice. BRAF and KRAS mutations are identified relatively frequently (6.1 and 4.9%, respectively) in GCB type DLBCL [25]. Although a cooperative effect between COMPASS components and RAS-RAF cascade mutations has yet to be reported, our results suggest that their potential combined impact on the pathogenesis of GCB DLBCL is worth investigating.
Development of plasma cell neoplasia in Utx insufficient Braf V600E mice
The most frequent neoplasm that developed in Utx insufficient BrafV600E mice was plasma cell neoplasm: 60%, 31.3%, and 35.3% in UtxΔ/YBrafV600E, UtxΔ/+BrafV600E, and UtxΔ/ΔBrafV600E mice, respectively (Fig. 1B right panel). These mice showed significant increase in the percentage of plasma cells in the BM and spleen (Fig. 3A) and many of them had splenomegaly and advanced anemia (Fig. 1C). Flow cytometric analysis and immunohistochemistry revealed the diffuse infiltration of CD138+ plasma cells in the BM, spleen, LNs, and peritoneal cavity (Fig. 3B left and middle panels and Supplementary Table 3). Genomic PCR analysis of purified plasma cells revealed that they were of clonal origin showing a monotonous pattern of rearrangement of the immunoglobulin heavy chain (Igh) gene (Fig. 3B upper right panel). M-spike was detected in serum protein electrophoresis (SPEP) assays in half of the mice with plasma cell neoplasms (Fig. 3B lower right panel and Supplementary Table 3). Some mice showed focal expansion of plasma cells, mimicking plasmacytoma (Fig. 3C). BM and spleen plasma cells from moribund mice showed typical plasma cell morphology (Fig. 3B middle panel, C, and D). These results indicate that the mice with plasma cell neoplasms recapitulate the major characteristics of human multiple myeloma (MM), however, most of the plasma cell neoplasms involved extramedullary organs (Supplementary Table 3).
A The percentage of plasma cells in the BM (left) and spleen (right) of mice that developed plasma cell neoplasms (BM, n = 23; spleen, n = 22), B-cell lymphoma (n = 11, each), LPD (n = 12, each), and Cre-negative control (n = 11, each). Bars indicate mean. **P < 0.01 by student t-test. B Left panel: representative plots of flow cytometric analysis of PI− BM and spleen cells from a 38-week-old Cre-negative male control and 25-week-old UtxΔ/yBrafV600E male mouse (BU-415). Proportions of plasma cells (CD138+B220–) are indicated. Middle panel: representative BM and spleen sections from BU-415 stained with hematoxylin and eosin or anti-CD138 antibody. Right panel: genomic PCR analysis of Igh rearrangement in BM plasma cells from a Cre-negative control and BU-415 (upper), and SPEP (Serum protein electrophoresis) performed on WT, BU333 (UtxΔ/+BrafV600E), and BU415 mice (lower). The positions of albumin and the different globulins are indicated. Arrowhead indicates an M-spike. C BM and spleen sections from BU-044 (UtxΔ/+BrafV600E) mouse stained with hematoxylin and eosin or anti-CD138 antibody. D Flow cytometric plot of PI− BM cells from BU-959 (UtxΔ/ΔBrafV600E) (left) and cytological features of BM cells from BU-959 observed by Wright-Giemsa staining (right). Arrowheads indicate malignant plasma cells.
We next performed transplantation assays. We purified CD138+ plasma cells by magnetic column-based purification (average purity 70%). We then transplanted them into CD45.1 (Ly5.1) congenic B57BL/6 mice or NOD.Cg-PrkdcscidIl2rgtm1Sug/Jic (NOG) immunocompromised mice via tail vein (Supplementary Table 4). We also performed transplantation using bulk cells from BM and spleen. Most of the mice developed lethal LPD-like disease, but only a fraction of recipient mice showed evident expansion of malignant plasma cells (Supplementary Table 4), suggesting that the engraftment and expansion of clonal plasma cells were less efficient than polyclonal Utx-null post-GC B-cells.
Modeling MM by using Utx insufficient myeloma cells
To perform detailed characterization of Utx insufficient plasma cell neoplasms, we established several cell lines from plasmacytic ascites. One cell line was obtained from a moribund UtxΔ/ΔBrafV600E mouse (BU-749, Supplementary Table 3) with an MM-like disease (Fig. 4A). The CD138+B220− malignant plasma cells grew on TSt-4 stromal cells [32] in the presence of interleukin-6 (IL-6), and were of clonal origin showing monoclonal Igl gene rearrangement (Fig. 4A right panel). These cells, designated as MM/BU749, successfully engrafted in sublethally irradiated NOG mice, which subsequently developed deadly disease (Fig. 4B). Mice with advanced disease showed paralysis of the hindlimbs and body weight loss (data not shown). Importantly, the secondary recipients developed lethal disease much earlier than the primary recipients (Fig. 4B).
A Left panel: representative plots of flow cytometric analysis of BM, mesenteric LN, and ascites from a 42-week-old UtxΔ/ΔBrafV600E female mouse (BU-749). Proportions of plasma cells (CD138+B220−PI−) are indicated. Right panel shows the characterization of an IL-6-dependent cell line derived from the plasmacytic ascites of BU-749. Cytological features observed by May-Giemsa staining (left), a representative plot of flow cytometric analysis of plasma cells (CD138+B220−PI−) (middle), and genomic PCR data of Igl Vκ-Jκ rearrangement (right) are depicted. Polyclonal spleen plasma cells (PCs) from control mice were used as a control of genomic PCR. B Upper panel: Schematic diagram of transplantation strategy. We injected 0.43 × 106 BU749 cells, which were described in (A) and cultured in the presence of IL-6, into 5 sublethally irradiated (1.5 Gy) NOG mice. We recovered BM cells from moribund recipient mice and injected 1 × 106 BM cells into secondary recipient NOG mice. Kaplan–Meier survival analysis of primary (n = 5) and secondary (n = 5) recipient mice is depicted (lower panel, left). Representative plots of flow cytometric analysis of BM from primary and secondary NOG recipient mice are shown (lower panel, right). Proportions of plasma cells (CD138+B220−PI−) are indicated. Statistical significance of survival difference was determined by the log-rank test. **P < 0.01. C A diagram summarizing 14 genes affected by somatic mutations detected in the indicated malignant plasma cells by whole exome sequencing. Non-recurrent missense mutations (139 genes) and the details of the samples are listed in Supplementary Table 5A–D.
To understand the collaborating somatic mutations acquired by Utx-insufficient BrafV600E plasma cell clones during disease initiation and progression, we performed whole exome sequencing (WES) using DNA of plasma cells purified from BM, spleen, and ascitic fluid of mice with plasma cell neoplasms and MM/BU749 cell line. DNA of corresponding tail or PB B cells was used as a control (Supplementary Table 5A). The mean depth of WES was 50X and the mean coverage of target sequences with an average depth of more than 10X was greater than 95% (Supplementary Tables 5A and B). A total of 153 somatic mutations were identified in 112 genes in 8 samples (Fig. 4C and Supplementary Tables 5C and D). Missense mutations were recurrently detected in at least 2 cases, in only three genes: E2f1, Muc4, and Vmn2r114. Other mutations included 5 nonsense mutations, 5 frameshift indels, 1 splice-site mutation, and 139 missense mutations (Fig. 4C and Supplementary Tables 5C and D). Unexpectedly, none of these mutations were major driver mutations in MM [16].
UTY is the Y chromosome homolog of UTX with weaker tumor-suppressive activity compared to UTX [33]. It has been reported that Uty expression protects Utx-/Y male mice from leukemogenesis [15]. In addition, UTY is frequently lost or mutated in pancreatic tumors with squamous differentiation in male patients [34]. Genomic loss of UTY in male cancer cell lines with inactivating UTX mutations (13/16, 81%) is significantly more frequent than UTY loss in UTX wild-type cancers (153/307, 49%) [18]. These findings support a tumor suppressive function of UTY in a setting of UTX insufficiency. However, our WES for male mice that developed MM-like disease did not show any abnormalities in Uty or Y-chromosome. Of interest, the mutation profiles of splenic and ascitic plasma cells from BU749 mouse were totally different, indicating the clonal heterogeneity of malignant plasma cells in the same mouse. These results indicate that Utx loss and BrafV600E are the main drivers of tumorigenesis in this MM model.
Catalytic activity is dispensable for the tumor suppressor function of UTX in MM
Making use of our Utx-deficient plasma cell line, we evaluated the impact of UTX add-back on Utx-null MM/BU749 cell growth. We conditionally overexpressed human UTX in MM/BU749 cells using a Tet-on lentivirus system. Exogenous UTX significantly impaired the proliferation of MM/BU749 in suspension culture in the presence of doxycycline (DOX) (Fig. 5A upper left panel). Growth suppressive effects of UTX add-back correlated well with the expression levels of exogenous UTX (Fig. 5A upper right panel). We next transplanted MM/BU749 cells with DOX-regulated exogenous UTX into NOG mice. In this experiment, we used NOD.Cg-PrkdcscidIl2rgtm1Sug Tg (CMV-IL6)/Jic (NOG-hIL-6) mice [24] to facilitate plasma cell engraftment. Induction of UTX in MM/BU749 clearly prevented the development of lethal disease in recipient mice and prolonged their survival (Fig. 5A lower panels), confirming the deleterious effects of UTX add-back on Utx-null plasma cell expansion in vivo. UTX add-back in UTX-null human MM cell line (RPMI8226) also impaired their growth (Fig. 5B left panel) and prolonged the survival of recipient mice in xenotransplantation assays (Fig. 5B right panel) as previously reported [19].
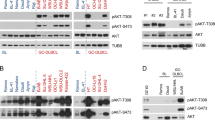
A Effects of UTX add-back on Utx-null BU749 cell growth. Upper left panel: Growth of BU749 with DOX-regulated human UTX in the presence and absence of DOX (2 μg/ml). Upper right panel: Expression of Flag-UTX protein was detected by western blotting with anti-Flag antibody. α-tubulin served as a loading control. Lower left panel: schematic diagram of transplantation strategy. Lower right panel: Kaplan–Meier survival analysis of sublethally (1.5 Gy) irradiated NOG-hIL-6 mice (n = 6, each) receiving 2 million BU749 cells per mouse. DOX was given to the mice in drinking water from day 40 post-transplantation and the mice were observed for 30 days. DOX dose: 0.2% (2 mg/mL) in drinking water, containing 3.5% sucrose, to be changed twice a week and protected from light. B Effects of UTX add-back on UTX-null human MM RPMI8226 cell growth. Left panel: growth of UTX-null human RPMI8226 cells transduced with the indicated lentiviruses. Middle panel: expression of Flag-UTX protein was detected by western blotting with anti-UTX and Flag antibodies. GAPDH served as a loading control. Right panel: Kaplan–Meier survival analysis of sublethally (1.5 Gy) irradiated NOG-hIL-6 mice receiving 3 million RPMI8226 cells transduced with either control (n = 8) or UTX (n = 8) lentivirus per mouse. C Upper panel: schematic representation of WT and demethylase inactive mutant UTX. Lower panel: effects of WT and demethylase inactive mutant UTX add-back on UTX-null human MM RPMI8226 cell growth. D Western blot analyses of global histone modification levels in plasma cells from control and UtxΔ/Δ mice using anti-H3K27me3, H3K27ac, and histone H3 antibodies. Levels of H3K27me3 and H3K27ac were normalized to the amount of H3 and are indicated relative to wild-type control values at the bottom. E Scatterplot showing the relationship of the fold enrichment values of H3K27me3 ChIP signals against the input signals (ChIP/input) at TSS ± 2.0 kb of RefSeq genes between BrafV600E and UtxΔ/ΔBrafV600E mice. The dotted light gray lines represent the boundaries for twofold increase and twofold decrease, respectively. The genes that gained and lost H3K27me3 levels greater than twofold in UtxΔ/ΔBrafV600E plasma cells compared with BrafV600E plasma cells are indicated in orange and blue, respectively. Cell growth data are shown as the mean ± SD of triplicate cultures. Statistical significance of survival difference and cell growth was determined by the log-rank test and the student t-test, respectively. *P < 0.05; **P < 0.01; ***P < 0.001.
To investigate the role of the demethylase activity of UTX in MM, we transduced RPMI8226 cells with WT and a demethylase-inactive mutant UTX. Point mutations at amino acid residues of the Fe(II)-binding motif in the JmjC domain (H1146 and E1148) abolish the demethylase activity of UTX (Fig. 5C upper panel) [35, 36]. Of note, add-back of H1146A/E1148A mutant impaired the growth of RPMI8226 cells in a manner similar to WT UTX (Fig. 5C lower panel). We then performed western blot analysis of histone modifications in plasma cells from UtxΔ/Δ mice and found no significant changes in both H3K27me3 and H3K27ac levels (Fig. 5D). In addition, ChIP-seq analysis did not reveal any significant change in H3K27me3 levels in UtxΔ/ΔBrafV600E plasma cells compared to BrafV600E plasma cells (Fig. 5E). These results indicate that UTX exerts a tumor suppressor function in a demethylase activity-independent manner.
Recently, it has been proposed that the tumor suppressor function of UTX largely relies on UTX condensation, higher-order assembles that are mediated by a core intrinsically disordered region (cIDR) [33]. We therefore tested the add-back of ΔcIDR UTX mutant in UTX-null MM cells. A UTX mutant lacking cIDR mostly failed to suppress the growth of the cells (Supplementary Fig. 2A, B). These results suggest that the cIDR domain is largely responsible for the tumor suppressor function of UTX in MM.
Utx loss with Braf V600E induces myeloma-like gene signature in plasma cells
To understand the impact of UTX loss on the plasma cell transcriptome, we performed RNA-seq of plasma cells from young (20 ~ 25-week-old) and old (40 ~ 50-week-old) Cγ1-Cre negative control mice, BrafV600E mice (31-week-old), young UtxΔ/ΔBrafV600E mice (20 ~ 25-week-old), Utx/BrafV600E mice with overt MM (MM1-3, 20 ~ 50-week-old), and BU749 (UtxΔ/ΔBrafV600E) cells. A Uniform Manifold Approximation and Projection (UMAP) analysis revealed that Utx loss together with BrafV600E readily induced moderate transcriptomic reprogramming of plasma cells towards MM-like transcriptome (Fig. 6A). Gene set enrichment analysis revealed activation of gene sets associated with cell cycle, Myc targets, and ribosome biogenesis and downregulation of gene sets associated with B lymphocytes and CD40 signal in Utx/BrafV600E mice (Fig. 6B). The change in the expression levels of these gene sets was moderate in UtxΔ/ΔBrafV600E mice without overt MM compared with those in Utx/BrafV600E mice with overt MM (Supplementary Fig. 3, Supplementary Tables 6A and B). We then defined differentially expressed genes (DEGs) between young UtxΔ/ΔBrafV600E, MM, and control plasma cells (FDR q < 0.01, Supplementary Table 7). Utx/BrafV600E plasma cells before and after overt MM showed gradual up-regulation and down-regulation of DEGs (Fig. 6C upper panel), including representative myeloma signature genes with upregulation of Myc, Ccnd2, E2f3, and Irf4 and downregulation of Cd19 and Ikzf3 (Fig. 6C lower panel, D). MYC is one of the key genes in the pathogenesis of multiple myeloma. Of interest, neither BrafV600E nor UtxΔ/Δ mice showed Myc activation in plasma cells. Intriguingly, UtxΔ/ΔBrafV600E mice before overt MM showed very mild albeit significant up-regulation of Myc compared to a mouse with overt MM (Fig. 6E). These results indicate that the myeloma-like transcriptional landscape develops gradually in Utx/BrafV600E plasma cells leading to growth advantage in vivo.
A UMAP plot based on the z-scores of expression values (DESeq2 normalized counts) of RNA-seq data of plasma cells from young (21 ~ 22-week-old) and old (40 ~ 55-week-old) control, BrafV600E (31-week-old), young UtxΔ/ΔBrafV600E (20-week-old) mice, compound mice with overt MM (MM1-3, 25 ~ 52-week-old), and BU749 (UtxΔ/ΔBrafV600E) cells. B GSEA using RNA-seq data of BM plasma cells from the indicated mice. Summary of GSEA data of representative gene sets is shown. Normalized enrichment scores (NES), nominal p values (NOM), and false discovery rates (FDR) are indicated. The GSEA data are listed in Supplementary Table 6A and B. C Upper panel; heatmap showing the z-scores of expression values (DESeq2 normalized counts) of DEGs (FDR q < 0.01) in BM plasma cells from compound mice with overt MM, BrafV600E and young UtxΔ/ΔBrafV600E mice in comparison with control plasma cells. Lower panel: volcano plots of DEGs normalized by DESeq2. Genes up-regulated and down-regulated are shown in red and blue, respectively. D Heatmap showing the z-scores of expression values (DESeq2 normalized counts) of representative myeloma signature genes. E Quantitative RT-PCR analysis of Myc in BM plasma cells from control, BrafV600E, UtxΔ/Δ, young UtxΔ/ΔBrafV600E, and MM UtxΔ/+BrafV600E mice. Actb was used to normalize the amount of input RNA. Data are shown as the mean ± SEM of triplicates. ***P < 0.001 by student t-test.
Characteristics of chromatin accessibility in Utx-null myeloma cells
To understand the transcriptional networks operating in Utx-null myeloma cells, we performed ATAC-seq in plasma cells from young Cγ1-Cre negative control, BrafV600E and UtxΔ/Δ mice (24-27-week-old), and BU749 (UtxΔ/ΔBrafV600E) plasma cells. UMAP analysis and hierarchical clustering of ATAC-seq data revealed that BU749 cells had largely different chromatin accessibility from control, BrafV600E, and UtxΔ/Δ plasma cells, while the changes among control, BrafV600E, and UtxΔ/Δ plasma cells were mild (Fig. 7A). We next defined differentially accessible regions (DARs) between control plasma cells and BU749 cells using DEseq2 and biological duplicates (n = 2). We found that 1422 and 907 DARs were open and closed in BU749 cells (p < 10−10, log2FC (fold change) >1), respectively (Fig. 7B, Supplementary Table 8). The majority of DARs were localized at the intergenic or intron regions that generally represent active or poised enhancers, or promoter regions. (Supplementary Fig. 4A). However, only a few DARs were detected that matched the strict criteria of p < 10−10, log2FC > 1 in BrafV600E and UtxΔ/Δ plasma cells (Fig. 7B).
A Left panel: UMAP plot based on ATAC peak profiling of plasma cells from young control, BrafV600E, UtxΔ/Δ mice (24–27-week-old), and BU749 (UtxΔ/ΔBrafV600E) plasma cells. Right panel: hierarchical clustering based on the Pearson’s correlation coefficient of ATAC peaks. B Volcano plots of DARs in BrafV600E and UtxΔ/Δ plasma cells and BU749 cells compared with young control plasma cells (p < 10−10, log2FC > 1). C Heat map showing the enrichment of the transcription factor binding motifs at open DARs in BU749 cells using hypergeometric distribution of Homer software. The combined peaks detected in all samples were used for the background to remove imbalance in the sequence content. D Heat map showing the enrichment of the indicated transcription factor binding motifs at each open DAR in BU749 cells calculated as in (C).
We then compared the frequencies of the transcription factor binding motifs at open DARs in BU749 cells using hypergeometric distribution of Homer software and calculated the p-values of enrichment. The combined peaks detected in all samples were used for the background to remove imbalance in the sequence content. This motif analysis revealed that the binding motifs of PU.1-IRF, PU.1-IRF8, PU.1, and SpiB were highly enriched in open DARs in BU749 cells (Fig. 7C and Supplementary Fig. 4B). PU.1-IRF motif was most frequently identified in open DARs in BU749 compared with control plasma cells (Fig. 7D). Importantly, Irf4 was one of the Irf genes with high expression in MM mice cells. In addition, Spi1 encoding Pu.1 was also highly expressed in MM cells (Supplementary Fig. 4C). The transcription factor IRF4 (interferon regulatory factor 4) is required for the development, maintenance, and function of plasma cells [37,38,39]. IRF4 is also a key regulator of multiple myeloma and directly activates MYC in myeloma cells [40]. IRF4 binds with low affinity to interferon sequence response elements (ISREs). In contrast, it binds with high affinity to ETS-IRF composite motifs (EICE) through interaction with the transcription factors PU.1 or SpiB [41, 42]. In addition, IRF4 also cooperatively assembles with BATF on composite AP-1-IRF (AICE) motifs [43]. To assess the correlation between changes in chromatin accessibility and transcription, DARs were connected to the nearest genes based on their distance to TSSs. The genes linked to open DARs, including those linked to DARs with PU.1-IRF motif, showed significantly higher levels of expression in BU749 cells than control plasma cells (Supplementary Fig. 4D).
To understand the epigenomic status of DARs, we profiled the acetylation status of H3K27 (H3K27ac) in plasma cells from control, BrafV600E, UtxΔ/Δ, and UtxΔ/ΔBrafV600E mice and BU749 (UtxΔ/ΔBrafV600E) plasma cells. The H3K27ac status followed a similar pattern to the chromatin accessibility profile, only a few differential H3K27ac peaks were detected in BrafV600E, UtxΔ/Δ, and UtxΔ/ΔBrafV600E plasma cells compared to control (Fig. 8A). In contrast, we found that the read counts of 1,352 and 3,252 peaks were increased and decreased in BU749 cells (q < 0.05), respectively (Fig. 8B, Supplementary Table 9). The majority of differential H3K27ac peaks were localized at the intergenic or intron regions that generally represent active or poised enhancers, or promoter regions. (Fig. 8C). The status of H3K27ac modification was well correlated with that of chromatin accessibility, particularly in the intergenic regions (Fig. 8D).
A Number of differential H3K27ac peaks in BrafV600E, UtxΔ/Δ, and UtxΔ/ΔBrafV600E plasma cells and BU749 cells compared to control. B Volcano plot of differential H3K27ac peaks in BU749 cells compared with control plasma cells (q < 0.05). C Percentage of each genomic annotation of H3K27ac peaks. D Correlation of H3K27ac and ATAC peaks in all genomic regions, promoter-TSS, and intergenic regions. P, p-value and R2, adjusted R-squared of the regression model. E Snapshots of ATAC-seq signals and H3K27ac modification at the Myc locus in control, BrafV600E, UtxΔ/Δ and UtxΔ/ΔBrafV600E plasma cells, and BU749 cells. The localization of PU.1-IRF motifs is indicated. The ChIP peaks of PU.1 and IRF4 in plasmablasts [44] are also depicted.
We then compared open DARs in BU749 cells with the published ChIP-seq data of IRF4 and PU.1 in plasmablasts [44]. As expected, many of the open DARs with PU.1-IRF motif overlapped with the IRF4 and PU.1 ChIP peaks in plasmablasts (Supplementary Fig. 4E). PU.1-IRF motif was identified at the Myc locus (Fig. 8E), a direct target of IRF4 [40]. However, chromatin accessibility at the Myc locus appeared to be already open with the presence of H3K27 acetylation in control plasma cells and showed no significant changes in UtxΔ/Δ plasma cells (Fig. 8E). This was in line with the insignificant change in Myc expression observed in UtxΔ/Δ plasma cells (Fig. 6E). In contrast, BU749 cells showed increased chromatin accessibility with the presence of H3K27 acetylation, particularly at +13 kb of the Myc locus that contained PU.1 and IRF4 binding sites (Fig. 8E). These data suggest that epigenomic reprogramming gradually proceeds in the absence of UTX and plasma cell clones that acquire myeloma-like properties are selected over time.
Sensitivity of UTX insufficient cells to pharmacological inhibition
UTX inactivating lesions in MM were associated with adverse overall survival, suggesting the association of UTX insufficiency with resistance to conventional therapy [17]. We therefore investigated the susceptibility of UTX insufficient myeloma cell lines to a proteasome inhibitor, bortezomib (BTZ); an immunomodulatory drug, lenalidomide; and a bromodomain inhibitor, JQ1. UTX insufficient cell lines (RPMI8226 and U266) required nearly a twofold higher concentration of BTZ to inhibit cell viability by 50% (cytotoxic concentration; CC50). Of note, UTX insufficient cell lines were highly resistant to lenalidomide and JQ1 compared with MM cell lines with WT UTX (Supplementary Fig. 5).
Discussion
In this study, we investigated the impact of conditional deletion of Utx in germinal center and post-germinal center B cells and plasma cells. Conditional deletion of Utx in a BrafV600E setting induced mature B cell malignancies including B cell lymphomas and multiple myeloma with the shortest survival being observed in UtxΔ/ΔBrafV600E mice. Our mouse model recapitulated the cardinal features of plasma cell neoplasms such as increased percentage of plasma cells in the BM, anemia, and large M spikes in SPEP. The main limitation was the absence of myeloma-like lytic bone lesions. Importantly, we did not find any major MM driver gene mutations or mutations in Uty in MM cells from moribund mice by WES. This well corresponded to the murine UtxΔ/Δ acute myeloid leukemia data that showed no recurrently mutated genes, with the exception of Skint11 (two of seven samples) [15]. These findings indicate that UTX insufficiency is a strong driver of myeloma development. Re-expression of UTX in both our Utx-deficient murine cell line (BU749) and human UTX-null RPMI8226 cells significantly inhibited the growth of the cells in vitro and in vivo. This is in agreement with the observations in human MM cell lines by Ezponda et al. [19]. Importantly, an enzymatically inactive UTX variant also suppressed the growth of UTX-null human RPMI8226 cells and this together with the insignificant change in H3K27me3 in UtxΔ/ΔBrafV600E plasma cells compared to control by ChIP-seq, suggests that the demethylase activity is dispensable for the tumor suppressor function of UTX in MM. While the H3K27-demethylase activity of UTX is thought to be essential for its tumor suppressor role in T-ALL [13, 14], it is deemed to be redundant in acute myeloid leukemia and pancreatic cancer [15, 34]. Recently, Shi et al. [33] reported that the tumor suppressor function of UTX largely relies on the TPR domain and the ability to form condensates via cIDR. Our results confirmed the essential role of the cIDR domain in the tumor suppressor activity of UTX in MM cells. In addition to the UTX inactivating mutations/deletions found in MM, inactivating mutations have also been detected in the components of COMPASS and its partner HATs, including KMT2C, CREBBP, and EP300 [16], further implicating dysregulated COMPASS activity in MM. Interestingly, the cooperativity between Utx loss and BrafV600E mutation in our mouse model points out the probable synergism between mutations in the COMPASS components and activation of the RAS-RAF-MEK-ERK/MAPK cascade in MM.
UTY has weaker tumor-suppressive activity than UTX. However, it functions as a tumor suppressor in a setting of UTX insufficiency [15, 18, 34]. Our WES for male MM mice did not show any abnormalities in Uty or Y-chromosome. The residual Uty function in UtxΔ/YBrafV600E males may explain the histological bias observed between male and female compound mice and the fact that UtxΔ/ΔBrafV600E females developed disease earlier than UtxΔ/YBrafV600E males. It would be intriguing to explore the role of Uty in this mouse model.
Our RNA-seq analysis revealed that the oncogene Myc was dramatically upregulated in UtxΔ/ΔBrafV600E MM cells but not in UtxΔ/Δ and UtxΔ/ΔBrafV600E plasma cells at early time points post-Utx deletion. Activation of MYC is common in MM [45] and overexpression of Myc in late B cells induced MM-like disease in mice [46]. In addition, Irf4, which is essential for the survival of MM cells [40], was also upregulated in mice with MM-like disease. Importantly, an autoregulatory loop between IRF4 and MYC has been described in MM [40]. Although GSEA revealed that gene sets related to Myc, multiple myeloma, and cell cycle were activated in young UtxΔ/ΔBrafV600E mice, the changes appeared to be very mild. Consistently, ATAC-seq and CUT&TAG data indicated very mild changes in chromatin accessibility and H3K27ac modification in UtxΔ/Δ plasma cells at an early time point post-Utx deletion. The PU.1-IRF motif most highly enriched in open DARs in UtxΔ/ΔBrafV600E MM cells was not enriched in open DARs in UtxΔ/Δ plasma cells. These results suggest that while UTX loss does not immediately induce drastic phenotypic changes in plasma cells, it allows them to undergo gradual genome-wide re-organization and transcriptional reprogramming. Subsequently, plasma cell clones that acquired MM-like properties are selected over time. This process could involve re-organization of epigenetic regulators such as histone modifiers, as we observed changes in chromatin accessibility and H3K27ac at the Myc locus. Alternatively, it is possible that UTX loss induces severe phenotypic changes in selected plasma cells, which expand to overt MM over time. Further analyses are needed to clarify the mechanism of myelomagenesis in our mouse model.
MM patients with a UTX deletion or mutation have a worse overall survival (OS) compared with those with WT UTX [17]. We confirmed that UTX insufficient cell lines were highly resistant to lenalidomide and JQ1 compared with MM cell lines with WT UTX. Bromodomain inhibitors negatively regulates promoter and enhancer activity, including those of well-known oncogenic transcription factor genes such as MYC and FOSL1 [47,48,49]. This suggests that UTX insufficiency induces enhancer reorganization at critical oncogenic gene loci and stabilizes their active enhanceosome. This is consistent with the marked activation of MYC in UTX insufficient myeloma cells in our MM mouse model.
In summary, we established a novel myeloma mouse model in which Utx loss and BrafV600E are combined. This study clearly demonstrates a catalytic activity-independent tumor suppressor function of UTX in MM and implicates its insufficiency in the transcriptional reprogramming of plasma cells. Our mouse model could be a useful tool for understanding the role of epigenetic dysfunction in mature B cell malignancies and studying novel therapeutic agents for MM.
Data availability
RNA- and ChIP-sequencing data and CUT&TAG data obtained in this study were deposited in DNA Data Bank of Japan (DDBJ) (accession numbers DRA13765 and DRA015180). A complete and detailed description of methods is provided in Supplementary Methods.
Change history
18 July 2023
A Correction to this paper has been published: https://doi.org/10.1038/s41375-023-01969-y
References
Michalak EM, Burr ML, Bannister AJ, Dawson MA. The roles of DNA, RNA and histone methylation in ageing and cancer. Nat Rev Mol Cell Biol. 2019;20:573–89.
Ntziachristos P, Abdel-Wahab O, Aifantis I. Emerging concepts of epigenetic dysregulation in hematological malignancies. Nat Immunol. 2016;17:1016–24.
Iwama A. Polycomb repressive complexes in hematological malignancies. Blood. 2017;130:23–29.
Laugesen A, Højfeldt JW, Helin K. Molecular mechanisms directing PRC2 recruitment and H3K27 methylation. Mol Cell. 2019;74:8–18.
Van der Meulen J, Speleman F, Van Vlierberghe P. The H3K27me3 demethylase UTX in normal development and disease. Epigenetics. 2014;9:658–68.
Mansour AA, Gafni O, Weinberger L, Zviran A, Ayyash M, Rais Y, et al. The H3K27 demethylase Utx regulates somatic and germ cell epigenetic reprogramming. Nature. 2012;488:409–13.
Shpargel KB, Sengoku T, Yokoyama S, Magnuson T. UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLOS Genet. 2012;8:e1002964.
Wang C, Lee J-E, Cho Y-W, Xiao Y, Jin Q, Liu C, et al. UTX regulates mesoderm differentiation of embryonic stem cells independent of H3K27 demethylase activity. Proc Natl Acad Sci USA. 2012;109:15324.
Morales Torres C, Laugesen A, Helin K. Utx is required for proper induction of ectoderm and mesoderm during differentiation of embryonic stem cells. PLoS One. 2013;8:e60020.
Sze CC, Shilatifard A. MLL3/MLL4/COMPASS family on epigenetic regulation of enhancer function and cancer. Cold Spring Harb Perspect Med. 2016;6:a026427.
Wang S-P, Tang Z, Chen C-W, Shimada M, Koche RP, Wang L-H, et al. A UTX-MLL4-p300 transcriptional regulatory network coordinately shapes active enhancer landscapes for eliciting transcription. Mol Cell. 2017;67:308–321.e306.
Wang L, Shilatifard A. UTX mutations in human cancer. Cancer Cell. 2019;35:168–76.
Ntziachristos P, Tsirigos A, Welstead GG, Trimarchi T, Bakogianni S, Xu L, et al. Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature. 2014;514:513–7.
Van der Meulen J, Sanghvi V, Mavrakis K, Durinck K, Fang F, Matthijssens F, et al. The H3K27me3 demethylase UTX is a gender-specific tumor suppressor in T-cell acute lymphoblastic leukemia. Blood. 2015;125:13–21.
Gozdecka M, Meduri E, Mazan M, Tzelepis K, Dudek M, Knights AJ, et al. UTX-mediated enhancer and chromatin remodeling suppresses myeloid leukemogenesis through noncatalytic inverse regulation of ETS and GATA programs. Nat Genet. 2018;50:883–94.
Walker BA, Mavrommatis K, Wardell CP, Ashby TC, Bauer M, Davies FE, et al. Identification of novel mutational drivers reveals oncogene dependencies in multiple myeloma. Blood. 2018;132:587.
Pawlyn C, Kaiser MF, Heuck C, Melchor L, Wardell CP, Murison A, et al. The spectrum and clinical impact of epigenetic modifier mutations in myeloma. Clin Cancer Res. 2016;22:5783–94.
van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41(May):521–3.
Ezponda T, Dupere-Richer D, Will CM, Small EC, Varghese N, Patel T, et al. UTX/KDM6A loss enhances the malignant phenotype of multiple myeloma and sensitizes cells to EZH2 inhibition. Cell Rep. 2017;21:628–40.
Corre J, Cleynen A, Robiou du Pont S, Buisson L, Bolli N, Attal M, et al. Multiple myeloma clonal evolution in homogeneously treated patients. Leukemia. 2018;32:2636–47.
Mercer K, Giblett S, Green S, Lloyd D, DaRocha Dias S, Plumb M, et al. Expression of endogenous oncogenic V600EB-raf induces proliferation and developmental defects in mice and transformation of primary fibroblasts. Cancer Res. 2005;65:11493–11500.
Sera Y, Nakata Y, Ueda T, Yamasaki N, Koide S, Kobayashi H, et al. UTX maintains the functional integrity of the murine hematopoietic system by globally regulating aging-associated genes. Blood. 2021;137:908–22.
Casola S, Cattoretti G, Uyttersprot N, Koralov SB, Seagal J, Hao Z, et al. Tracking germinal center B cells expressing germ-line immunoglobulin gamma1 transcripts by conditional gene targeting. Proc Natl Acad Sci USA. 2006;103:7396–401.
Hanazawa A, Ito R, Katano I, Kawai K, Goto M, Suemizu H, et al. Generation of Human Immunosuppressive Myeloid Cell Populations in Human Interleukin-6 Transgenic NOG Mice. Front Immunol. 2018;9:152.
Schmitz R, Wright GW, Huang DW, Johnson CA, Phelan JD, Wang JQ, et al. Genetics and Pathogenesis of Diffuse Large B-Cell Lymphoma. N. Engl J Med. 2018;378:1396–407.
Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303.
Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, Trifonov V, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471:189–95.
Ortega-Molina A, Boss IW, Canela A, Pan H, Jiang Y, Zhao C, et al. The histone lysine methyltransferase KMT2D sustains a gene expression program that represses B cell lymphoma development. Nat Med. 2015;21:1199–208.
Kung AL, Rebel VI, Bronson RT, Ch’ng L-E, Sieff CA, Livingston DM, et al. Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes Dev. 2000;14:272–7.
Xu W, Fukuyama T, Ney PA, Wang D, Rehg J, Boyd K, et al. Global transcriptional coactivators CREB-binding protein and p300 are highly essential collectively but not individually in peripheral B cells. Blood. 2006;107:4407–16.
Li X, Zhang Y, Zheng L, Liu M, Chen CD, Jiang H. UTX is an escape from X-inactivation tumor-suppressor in B cell lymphoma. Nat Commun. 2018;9:2720.
Masuda K, Kubagawa H, Ikawa T, Chen C-C, Kakugawa K, Hattori M, et al. Prethymic T-cell development defined by the expression of paired immunoglobulin-like receptors. EMBO J. 2005;24:4052–60.
Shi B, Li W, Song Y, Wang Z, Ju R, Ulman A, et al. UTX condensation underlies its tumour-suppressive activity. Nature. 2021;597(Sep):726–31.
Andricovich J, Perkail S, Kai Y, Casasanta N, Peng W, Tzatsos A. Loss of KDM6A Activates super-enhancers to induce gender-specific squamous-like pancreatic cancer and confers sensitivity to BET inhibitors. Cancer Cell. 2018;33:512–526.e518.
Hong S, Cho Y-W, Yu L-R, Yu H, Veenstra TD, Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci USA. 2007;104:18439–44.
Sengoku T, Yokoyama S. Structural basis for histone H3 Lys 27 demethylation by UTX/KDM6A. Genes Dev. 2011;25:2266–77.
Ochiai K, Maienschein-Cline M, Simonetti G, Chen J, Rosenthal R, Brink R, et al. Transcriptional regulation of germinal center B and plasma cell fates by dynamical control of IRF4. Immunity. 2013;38:918–29.
Sciammas R, Shaffer AL, Schatz JH, Zhao H, Staudt LM, Singh H. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 2006;25:225–36.
Tellier J, Shi W, Minnich M, Liao Y, Crawford S, Smyth GK, et al. Blimp-1 controls plasma cell function through the regulation of immunoglobulin secretion and the unfolded protein response. Nat Immunol. 2016;17:323–30.
Shaffer AL, Emre NCT, Lamy L, Ngo VN, Wright G, Xiao W, et al. IRF4 addiction in multiple myeloma. Nature. 2008;454:226–31.
Brass AL, Zhu AQ, Singh H. Assembly requirements of PU.1–Pip (IRF-4) activator complexes: inhibiting function in vivo using fused dimers. EMBO J. 1999;18:977–91.
Eisenbeis CF, Singh H, Storb U. Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes Dev. 1995;9:1377–87.
Glasmacher E, Agrawal S, Chang AB, Murphy TL, Zeng W, Vander Lugt B, et al. A genomic regulatory element that directs assembly and function of immune-specific AP-1-IRF complexes. Science. 2012;338:975–80.
Minnich M, Tagoh H, Bönelt P, Axelsson E, Fischer M, Cebolla B, et al. Multifunctional role of the transcription factor Blimp-1 in coordinating plasma cell differentiation. Nat Immunol. 2016;17:331–43.
Morgan GJ, Walker BA, Davies FE. The genetic architecture of multiple myeloma. Nat Rev Cancer. 2012;12:335–48.
Chesi M, Robbiani DF, Sebag M, Chng WJ, Affer M, Tiedemann R, et al. AID-dependent activation of a MYC transgene induces multiple myeloma in a conditional mouse model of post-germinal center malignancies. Cancer Cell. 2008;13:167–80.
Delmore J, Issa G, Lemieux M, Rahl P, Shi J, Jacobs H, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–17.
Baker EK, Taylor S, Gupte A, Sharp PP, Walia M, Walsh NC, et al. BET inhibitors induce apoptosis through a MYC independent mechanism and synergise with CDK inhibitors to kill osteosarcoma cells. Sci Rep. 2015;5:10120.
Lockwood WW, Zejnullahu K, Bradner JE, Varmus H. Sensitivity of human lung adenocarcinoma cell lines to targeted inhibition of BET epigenetic signaling proteins. Proc Natl Acad Sci USA. 2012;109:19408.
Acknowledgements
We would like to thank Drs. Hao Jiang for providing us with the UTX mutant plasmids. We are grateful for Drs. Omar Abdel-Wahab, Yutaka Okuno, and Tohru Iseki for their advice and Yuko Yamagata and Makiko Miyota for their technical assistance. The super-computing resource was provided by the Human Genome Center, the Institute of Medical Science, the University of Tokyo. This work was supported in part by Grants-in-Aid for Scientific Research (#26860719, #16K09839, #19H05653, #19K08807, #22K08495, #19K21281, and #22K16318) and Scientific Research on Innovative Area “Replication of Non-Genomic Codes” (#19H05746) from Japanese Society for the Promotion of Science (JSPS), Japan, and Moonshot project (#21zf0127003h0001) from AMED, Japan, and grants from the Yasuda Medical Foundation, the Mochida Memorial Foundation, the Kanae Foundation for the promotion of Medical Science, the Takeda Science Foundation, the MSD Life Science Foundation, and the Chiba University SEEDS Fund (Chiba University Open Recruitment for International Exchange Program).
Funding
Open access funding provided by The University of Tokyo.
Author information
Authors and Affiliations
Contributions
OR, and NM performed the experiments, analyzed the results, made the figures, and actively wrote the manuscript; MO, SM, NT, SK, AS, YM-N, MR, NI, YN-T, KA, CW, AS, MS, MW, SY, TS, KY, YF, TC assisted with the experiments; ES, CN, J-IT, Y-TT, KCA interpreted data; HH developed and provided mice; and A.I. conceived of and directed the project, secured funding, and actively wrote the manuscript. All authors reviewed the manuscript and approved the final version for submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: The list of authors has been revised. The author Ryoji Ito was added.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rizq, O., Mimura, N., Oshima, M. et al. UTX inactivation in germinal center B cells promotes the development of multiple myeloma with extramedullary disease. Leukemia 37, 1895–1907 (2023). https://doi.org/10.1038/s41375-023-01928-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-023-01928-7