Abstract
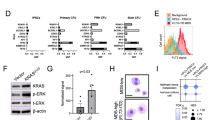
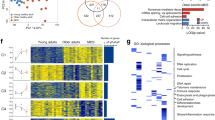
Primary myelofibrosis (PMF) is a hematopoietic stem cell (HSC) disease, characterized by aberrant differentiation of all myeloid lineages and profound disruption of the bone marrow niche. PMF samples carry several mutations, but their cell origin and hierarchy in regulating the different waves of clonal and aberrant myeloproliferation from the prime HSC compartment is poorly understood. Genotyping of >2000 colonies from CD133+HSC and progenitors from PMF patients confirmed the complex genetic heterogeneity within the neoplastic population. Notably, mutations in chromatin regulators ASXL1 and/or EZH2 were identified as the first genetic lesions, preceding both JAK2-V617F and CALR mutations, and are thus drivers of clonal myelopoiesis in a PMF subset. HSC from PMF patients with double ASXL1/EZH2 mutations exhibited significantly higher engraftment in immunodeficient mice than those from patients without histone modifier mutations. EZH2 mutations correlate with aberrant erythropoiesis in PMF patients, exemplified by impaired maturation and cell cycle arrest of erythroid progenitors. These data underscore the importance of post-transcriptional modifiers of histones in neoplastic stem cells, whose clonal growth sustains aberrant myelopoiesis and expansion of pre-leukemic clones in PMF.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Campbell P, Green A. The myeloproliferative disorders. New Engl J Med. 2006;355:2452–66.
Rumi E, Cazzola M. Diagnosis, risk stratification, and response evaluation in classical myeloproliferative neoplasms. Blood. 2017;129:680–92.
Nangalia J, Massie C, Baxter E, Nice F, Gundem G, Wedge D, et al. Somatic CALR mutations in myeloproliferative neoplasms with nonmutated JAK2. New Engl J Med. 2013;369:2391–405.
Rampal R, Al-Shahrour F, Abdel-Wahab O, Patel J, Brunel J-P, Mermel C, et al. Integrated genomic analysis illustrates the central role of JAK-STAT pathway activation in myeloproliferative neoplasm pathogenesis. Blood. 2014;123:e123–133.
Shih A, Abdel-Wahab O, Patel J, Levine R. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer. 2012;12:599–612.
Vainchenker W, Kralovics R. Genetic basis and molecular pathophysiology of classical myeloproliferative neoplasms. Blood. 2017;129:667–79.
Lundberg P, Karow A, Nienhold R, Looser R, Hao-Shen H, Nissen I, et al. Clonal evolution and clinical correlates of somatic mutations in myeloproliferative neoplasms. Blood. 2014;123:2220–8.
Ortmann CA, Kent DG, Nangalia J, Silber Y, Wedge DC, Grinfeld J, et al. Effect of mutation order on myeloproliferative neoplasms. New Engl J Med. 2015;372:601–12.
Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506:328–33.
Triviai I, Stübig T, Niebuhr B, Hussein K, Tsiftsoglou A, Fehse B, et al. CD133 marks a stem cell population that drives human primary myelofibrosis. Haematologica. 2015;100:768–79.
Thol F, Suchanek K, Koenecke C, Stadler M, Platzbecker U, Thiede C, et al. SETBP1 mutation analysis in 944 patients with MDS and AML. Leukemia. 2013;27:2072–5.
Thol F, Klesse S, Köhler L, Gabdoulline R, Kloos A, Liebich A, et al. Acute myeloid leukemia derived from lympho-myeloid clonal hematopoiesis. Leukemia. 2017;31:1286–95.
Badbaran A, Fehse B, Christopeit M, Aranyossy T, Ayuk F, Wolschke C, et al. Digital-PCR assay for screening and quantitative monitoring of calreticulin (CALR) type-2 positive patients with myelofibrosis following allogeneic stem cell transplantation. Bone Marrow Transplant. 2016;51:872–3.
Kröger N, Badbaran A, Holler E, Hahn J, Kobbe G, Bornhäuser M, et al. Monitoring of the JAK2-V617F mutation by highly sensitive quantitative real-time PCR after allogeneic stem cell transplantation in patients with myelofibrosis. Blood. 2007;109:1316–21.
Alchalby H, Badbaran A, Bock O, Fehse B, Bacher U, Zander A, et al. Screening and monitoring of MPL W515L mutation with real-time PCR in patients with myelofibrosis undergoing allogeneic-SCT. Bone Marrow Transplant. 2010;45:1404–7.
Abdel-Wahab O, Adli M, LaFave LM, Gao J, Hricik T, Shih AH, et al. ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell. 2012;22:180–93.
Dey A, Seshasayee D, Noubade R, French D, Liu J, Chaurushiya M, et al. Loss of the tumor suppressor BAP1 causes myeloid transformation. Science. 2012;337:1541–6.
Carbuccia N, Murati A, Trouplin V, Brecqueville M, Adélaïde J, Rey J, et al. Mutations of ASXL1 gene in myeloproliferative neoplasms. Leukemia. 2009;23:2183–6.
Sorigué M, Ribera J, García O, Cabezón M, Vélez P, Marcé S, et al. Highly variable mutational profile of ASXL1 in myelofibrosis. Eur J Haematol. 2016;97:331–5.
Guglielmelli P, Pacilli A, Rotunno G, Rumi E, Rosti V, Delaini F, et al. Presentation and outcome of patients with 2016 WHO diagnosis of prefibrotic and overt primary myelofibrosis. Blood. 2017;129:3227–36.
Anjos-Afonso F, Currie E, Palmer H, Foster K, Taussig D, Bonnet D. CD34(-) cells at the apex of the human hematopoietic stem cell hierarchy have distinctive cellular and molecular signatures. Cell Stem Cell. 2013;13:161–74.
Bhatia M, Bonnet D, Murdoch B, Gan O, Dick J. A newly discovered class of human hematopoietic cells with SCID-repopulating activity. Nat Med. 1998;4:1038–45.
Goodell M, Rosenzweig M, Kim H, Marks D, DeMaria M, Paradis G, et al. Dye efflux studies suggest that hematopoietic stem cells expressing low or undetectable levels of CD34 antigen exist in multiple species. Nat Med. 1997;3:1337–45.
Ishii M, Matsuoka Y, Sasaki Y, Nakatsuka R, Takahashi M, Nakamoto T, et al. Development of a high-resolution purification method for precise functional characterization of primitive human cord blood-derived CD34-negative SCID-repopulating cells. Exp Hematol. 2011;39:203–13.
Manz M, Milyamoto T, Akashi K, Weissmann I. Prospective isolation of human clonogenic common myeloid progenitors. Proc Natl Acad Sci USA. 2002;99:11872–7.
Nangalia J, Nice FL, Wedge DC, Godfrey AL, Grinfeld J, Thakker C, et al. DNMT3A mutations occur early or late in patients with myeloproliferative neoplasms and mutation order influences phenotype. Haematologica. 2015;100:42.
Steensma DP, Bejar R, Jaiswal S, Lindsley RC, Sekeres MA, Hasserjian RP, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126:9–16.
Zink F, Stacey SN, Norddahl GL, Frigge ML, Magnusson OT, Jonsdottir I, et al. Clonal hematopoiesis, with and without candidate driver mutations, is common in the elderly. Blood. 2017;130:742–52.
Busque L, Patel J, Figueroa M, Vasanthakumar A, Provost S, Hamilou Z, et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat Genet. 2012;44:1179–81.
Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Massé A, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360:2289–301.
Sashida G, Wang C, Tomioka T, Oshima M, Aoyama K, Kanai A, et al. The loss of Ezh2 drives the pathogenesis of myelofibrosis and sensitizes tumor-initiating cells to bromodomain inhibition. J Exp Med. 2016;213:1459–77.
Shi H, Yamamoto S, Sheng M, Bai J, Zhang P, Chen R, et al. ASXL1 plays an important role in erythropoiesis. Sci Rep. 2016;6:28789.
Shimizu T, Kubovcakova L, Nienhold R, Zmajkovic J, Meyer SC, Hao-Shen H, et al. Loss of Ezh2 synergizes with JAK2-V617F in initiating myeloproliferative neoplasms and promoting myelofibrosis. J Exp Med. 2016;213:1479–96.
Yang Y, Akada H, Nath D, Hutchison RE, Mohi G. Loss of Ezh2 cooperates with Jak2V617F in the development of myelofibrosis in a mouse model of myeloproliferative neoplasm. Blood. 2016;127:3410–23.
Acknowledgements
The authors would like to thank Laura Montanus, Marla Wobbe, Roman Kanke and Martin Wichmann for their contribution to PCR and sequencing experiments and the personnel of the Core Facility in University Medical Center Hamburg Eppendorf for their assistance in FACS experiments. We thank Dr. Robert Geffers and Dr. Michael Jarek from the Genome Analytics Group, Helmholtz Centre for Infection Research, Braunschweig, Germany for next-generation sequencing services.
This work was supported from grants of the European Hematology Association, the José Carreras Leukaemia Foundation and Else Kröner-Fresenius-Foundation to IT and grants from the German Federal Ministry of Education and Research grant 01EO0802 (IFB-Tx) and from DFG grants HE 5240/5-1 and HE 5240/6-1 to MH.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Triviai, I., Zeschke, S., Rentel, J. et al. ASXL1/EZH2 mutations promote clonal expansion of neoplastic HSC and impair erythropoiesis in PMF. Leukemia 33, 99–109 (2019). https://doi.org/10.1038/s41375-018-0159-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-018-0159-0
This article is cited by
-
Clonal selection of hematopoietic stem cells after gene therapy for sickle cell disease
Nature Medicine (2023)
-
Molecular pathogenesis of the myeloproliferative neoplasms
Journal of Hematology & Oncology (2021)
-
Philadelphia-Negative Myeloproliferative Neoplasms: Laboratory Workup in the Era of Next-Generation Sequencing
Current Hematologic Malignancy Reports (2019)
-
The ruxolitinib effect: understanding how molecular pathogenesis and epigenetic dysregulation impact therapeutic efficacy in myeloproliferative neoplasms
Journal of Translational Medicine (2018)