Abstract
Objective
To evaluate patterns of genetic testing among infants with CHD at a tertiary care center.
Study design
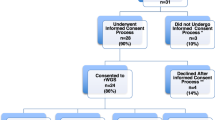
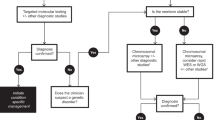
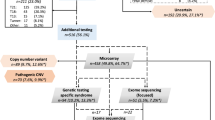
We conducted a retrospective observational cohort study of infants in the NICU with suspicion of a genetic disorder. 1075 of 7112 infants admitted to BCH had genetic evaluation including 329 with CHD and 746 without CHD. 284 of 525 infants with CHD admitted to CMHH had genetic evaluation. Patterns of testing and diagnoses were compared.
Results
The rate of diagnosis after testing was similar for infants with or without CHD (38% [121/318] vs. 36% [246/676], p = 0.14). In a multiple logistic regression, atrioventricular septal defects were most high associated with genetic diagnosis (odds ratio 29.99, 95% confidence interval 2.69–334.12, p < 0.001).
Conclusions
Infants with suspicion of a genetic disorder with CHD had similar rates of molecular diagnosis as those without CHD. These results support a role for genetic testing among NICU infants with CHD.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
Data availability
The datasets generated during and/or analysed during the current study are not publicly available due confidentiality but are available from the corresponding author on reasonable request.
References
Wu W, He J, Shao X. Incidence and mortality trend of congenital heart disease at the global, regional, and national level, 1990–2017. Medicine. 2020;99:e20593.
Giang KW, Mandalenakis Z, Fedchenko M, Eriksson P, Rosengren A, Norman M, et al. Congenital heart disease: changes in recorded birth prevalence and cardiac interventions over the past half-century in Sweden. Eur J Prev Cardiol. 2023;30:169–76.
Egbe A, Lee S, Ho D, Uppu S, Srivastava S. Prevalence of congenital anomalies in newborns with congenital heart disease diagnosis. Ann Pediatr Cardiol. 2014;7:86.
Zimmerman MS. Global, regional, and national burden of congenital heart disease, 1990–2017: a systematic analysis for the Global Burden of Disease study 2017. Lancet Child Adolesc Health. 2020;4:185–200.
Pierpont ME, Brueckner M, Chung WK, Garg V, Lacro RV, McGuire AL, et al. Genetic basis for congenital heart disease: revisited: a scientific statement from the American Heart Association. Circulation. 2018;138:e653–e711.
Morton SU, Quiat D, Seidman JG, Seidman CE. Genomic frontiers in congenital heart disease. Nat Rev Cardiol. 2022;19:26–42.
Peyvandi S, Lupo PJ, Garbarini J, Woyciechowski S, Edman S, Emanuel BS, et al. 22q11.2 deletions in patients with conotruncal defects: Data from 1,610 consecutive cases. Pediatr Cardiol. 2013;34:1687–94.
Vorstman JAS, Jalali GR, Rappaport AM, Scott C, Emanuel BS, et al. MLPA: a rapid, reliable, and sensitive method for detection and analysis of abnormalities of 22q. Hum Mutat. 2006;27:814–21.
Chaix MA. Genetic testing in congenital heart disease: a clinical approach. World J Cardiol. 2016;8:180.
Smith HS, Swint JM, Lalani SR, Otto MCO, Yamal J, Russell HV, et al. Exome sequencing compared with standard genetic tests for critically ill infants with suspected genetic conditions. Genet Med. 2020;22:1303–10.
Yang L, Wei Z, Chen X, Hu L, Peng X, Wang J, et al. Use of medical exome sequencing for identification of underlying genetic defects in <scp>NICU</scp>: experience in a cohort of 2303 neonates in China. Clin Genet. 2022;101:101–9.
Meng L, Pammi M, Saronwala A, Magoulas P, Ghazi AR, Vetrini F, et al. Use of exome sequencing for infants in intensive care units. JAMA Pediatr. 2017;171:e173438.
Wang H, Xiao F, Dong X, Yulan L, Cheng G, Wang L, et al. Diagnostic and clinical utility of next‐generation sequencing in children born with multiple congenital anomalies in the China neonatal genomes project. Hum Mutat. 2021;42:434–44.
Wu B, Kang W, Wang Y, Zhuang D, Chen L, Pan X, et al. Application of full-spectrum rapid clinical genome sequencing improves diagnostic rate and clinical outcomes in critically Ill infants in the China neonatal genomes project. Crit Care Med. 2021;49:1674–83.
Lunke S, Eggers S, Wilson M, Patel C, Barnett C, Pinner J, et al. Feasibility of ultra-rapid exome sequencing in critically Ill infants and children with suspected monogenic conditions in the Australian Public Health Care system. JAMA. 2020;323:2503.
Dimmock D, Caylor S, Waldman B, Benson W, Ashburner C, Carmichael J, et al. Project Baby Bear: rapid precision care incorporating rWGS in 5 California children’s hospitals demonstrates improved clinical outcomes and reduced costs of care. Am J Hum Genet. 2021;108:1231–8.
Bowling KM, Thompson ML, Finnila CR, Hiatt SM, Latner DR, Amaral MD, et al. Genome sequencing as a first-line diagnostic test for hospitalized infants. Genet Med. 2022;24:851–61.
D’Gama AM, Del Rosario MC, Bresnahan MA, Yu TW, Wojcik MH, Agrawal PB. Integrating rapid exome sequencing into NICU clinical care after a pilot research study. NPJ Genom Med. 2022;7:51.
Findley TO, Crain AK, Mahajan S, Deniwar A, Davis J, Zavala AS, et al. Congenital heart defects and copy number variants associated with neurodevelopmental impairment. Am J Med Genet A. 2022;188:13–23.
Elliott AM, Souich C, Lehman A, Guella I, Evans DM, Candido T, et al. RAPIDOMICS: rapid genome-wide sequencing in a neonatal intensive care unit—successes and challenges. Eur J Pediatr. 2019;178:1207–18.
Williamson SL, Rasanayagam CN, Glover KJ, Baptista J, Naik S, Satodia P, et al. Rapid exome sequencing: revolutionises the management of acutely unwell neonates. Eur J Pediatr. 2021;180:3587–91.
Shikany AR, Landis BJ, Parrott A, Miller EM, Coyan A, Walters L. et al. A Comprehensive clinical genetics approach to critical congenital heart disease in infancy. J Pediatr. 2020;227:231–238.e14.
Durbin MD, Helvaty LR, Li M, Ware SM. A multicenter cross-sectional study in infants with congenital heart defects demonstrates high diagnostic yield of genetic testing but variable evaluation practices. Genet Med Open. 2023;1:100814.
Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–64.
Gill K, Sasaki J, Jayakar P, Sosa L, Welch E. Chromosomal microarray detects genetic risks of neurodevelopmental disorders in newborns with congenital heart disease. Cardiol Young. 2021;31:1275–82.
Wu X, Li R, Fu F, Pan M, Han J, Yang X, et al. Chromosome microarray analysis in the investigation of children with congenital heart disease. BMC Pediatr. 2017;17:117.
Durbin MD, Fairman K, Helvaty LR, Huang M, Li M, Abreu D. et al. Genetic testing guidelines impact care in newborns with congenital heart defects. J Pediatr. 2023;260:113495.
Mone F, Eberhardt RY, Morris RK, Hurles ME, McMullan DJ, Maher ER, et al. COngenital heart disease and the diagnostic yield with exome sequencing (CODE) study: prospective cohort study and systematic review. Ultrasound Obstet Gynecol. 2021;57:43–51.
Paige SL, Saha P, Priest JR. Beyond gene panels: whole exome sequencing for diagnosis of congenital heart disease. Circ Genom Precis Med. 2018;11:e002097.
Acknowledgements
We would like to thank the infants, families, and providers in the BCH and UT NICUs.
Funding
SUM was supported by an American Heart Association Career Development Award and NIH/NHLBI R03HL150412 and K08HL157653. MW was supported by NIH/NICHD K23HD102589.
Author information
Authors and Affiliations
Contributions
TOF, PBA, MW and SUM conceptualized this work. EED, TOF, RH, ZSHK, RS, DS, AMD, HAF, MW and SUM participated in the acquisition and analysis of data. EED, TOF, RH, AMD, HAF, PBA, MW and SUM interpreted the data and results. EED, TOF, RH, ZSHD, AMD, PBA, MW and SUM drafted the manuscript. All authors critically reviewed the work for important intellectual content and approved the final version. Each author is accountable for their contribution to the manuscript, including related to the accuracy and integrity of the work.
Corresponding author
Ethics declarations
Competing interests
MW has received compensation for consultation to GeneDx, Illumina, and Sanofi. The other authors declare no competing financial interests in relation to the work described.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
D’Souza, E.E., Findley, T.O., Hu, R. et al. Genomic testing and molecular diagnosis among infants with congenital heart disease in the neonatal intensive care unit. J Perinatol (2024). https://doi.org/10.1038/s41372-024-01935-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41372-024-01935-1