Abstract
Objective
To determine detection rates of genetic disease in a level IV neonatal intensive care unit (NICU) and cost of care.
Study design
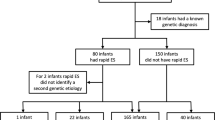
We divided 2703 neonates, admitted between 2013 and 2016 to a level IV NICU, into two epochs and determined how genetic testing utilization, genetic diagnoses identified, and cost of NICU care changed over time.
Result
The increasing use of multi-gene panels 104 vs 184 (P = 0.02) and whole exome sequencing (WES) 9 vs 28 (P = 0.03) improved detection of genetic disease, 9% vs 12% (P < 0.01). Individuals with genetic diagnoses had higher mean NICU charges, $723,422 vs $417,013 (P < 0.01) secondary to longer lengths of stay, not genetic services.
Conclusion
The increased utilization of broad genetic testing improved the detection of genetic disease but contributed minimally to the cost of care while bolstering understanding of the patient’s condition and prognosis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
De-identified data can be made available on an individual request.
References
Canfield MA, Honein MA, Yuskiv N, Xing J, Mai CT, Collins JS, et al. National estimates and race/ethnic-specific variation of selected birth defects in the United States, 1999–2001. Birth Defects Res. Part A, Clin Mol Teratol. 2006;76:747–56. https://doi.org/10.1002/bdra.20294.
Parker SE, Mai CT, Canfield MA, Rickard R, Wang Y, Meyer RE, et al. National birth defects prevention network. Updated National Birth Prevalence estimates for selected birth defects in the United States, 2004–2006. Birth Defects Res A Clin Mol Teratol. 2010;88:1008–16. https://doi.org/10.1002/bdra.20735.
Lu XY, Phung MT, Shaw CA, Pham K, Neil SE, Patel A, et al. Genomic imbalances in neonates with birth defects: high detection rates by using chromosomal microarray analysis. Pediatrics. 2008;122:1310–8. https://doi.org/10.1542/peds.2008-0297.
Blencowe H, Moorthie S, Petrou M, Hamamy H, Povey S, Bittles A, et al. Rare single gene disorders: estimating baseline prevalence and outcomes worldwide. J Community Genet. 2018;9:397–406. https://doi.org/10.1007/s12687-018-0376-2.
Swaggart KA, Swarr DT, Tolusso LK, He H, Dawson DB, Suhrie KR. Making a genetic diagnosis in a level IV neonatal intensive care unit population: who, when, how, and at what cost? J Pediatr. 2019;213:211–7. https://doi.org/10.1016/j.jpeds.2019.05.054.
Berg JS, Agrawal PB, Bailey DB, Beggs A, Brenner SE, Brower AM, et al. Newborn sequencing in genomic medicine and public health. Pediatrics. 2017;139:e20162252. https://doi.org/10.1542/peds.2016-2252.
Daoud H, Luco SM, Li R, Bareke E, Beaulieu C, Jarinova O, et al. Next-generation sequencing for diagnosis of rare diseases in the neonatal intensive care unit. CMAJ. 2016;188:E254–60. https://doi.org/10.1503/cmaj.150823.
Willig LK, Petrikin JE, Smith LD, Saunders CJ, Thiffault I, Miller NA, et al. Whole-genome sequencing for identification of Mendelian disorders in critically ill infants: a retrospective analysis of diagnostic and clinical findings. Lancet Respiratory Med. 2015;3:377–87. https://doi.org/10.1016/s2213-2600(15)00139-3.
Meng L, Pammi M, Saronwala A, Magoulas P, Ghazi AR, Vetrini F, et al. Use of exome sequencing for infants in intensive care units: ascertainment of severe single-gene disorders and effect on medical management. JAMA Pediatr. 2017;171:e173438. https://doi.org/10.1001/jamapediatrics.2017.3438.
Bamshad MJ, Nickerson DA, Chong JX. Mendelian gene discovery: fast and furious with no end in sight. Am J Hum Genet. 2019;105:448–55. https://doi.org/10.1016/j.ajhg.2019.07.011.
Maron JL, Kingsmore SF, Wigby K, Chowdhury S, Dimmock D, Poindexter B, et al. Novel variant findings and challenges associated with the clinical integration of genomic testing: an interim report of the genomic medicine for Ill neonates and infants (GEMINI) study. JAMA Pediatr. 2021;175:e205906. https://doi.org/10.1001/jamapediatrics.2020.5906.
Dimmock DP, Clark MM, Gaughran M, Cakici JA, Caylor SA, Clarke C, et al. An RCT of rapid genomic sequencing among seriously Ill infants results in high clinical utility, changes in management, and low perceived harm. Am J Hum Genet. 2020;107:942–52. https://doi.org/10.1016/j.ajhg.2020.10.003.
Zhu T, Gong X, Bei F, Ma L, Chen Y, Zhang Y, et al. Application of next-generation sequencing for genetic diagnosis in neonatal intensive care units: results of a multicenter study in China. Front Genet. 2020;11:1298. https://doi.org/10.3389/fgene.2020.565078.
Manickam K, McClain MR, Demmer LA, Biswas S, Kearney HM, Malinowski J, et al. Exome and genome sequencing for pediatric patients with congenital anomalies or intellectual disability: an evidence-based clinical guideline of the American College of Medical Genetics and Genomics (ACMG).
Persson M, Cnattingius S, Villamor E, Söderling J, Pasternak B, Stephansson O, et al. Risk of major congenital malformations in relation to maternal overweight and obesity severity: cohort study of 1.2 million singletons. BMJ. 2017;357:j2563. https://doi.org/10.1136/bmj.j2563.
Eldomery MK, Coban-Akdemir Z, Harel T, Rosenfeld JA, Gambin T, Stray-Pedersen A, et al. Lessons learned from additional research analyses of unsolved clinical exome cases. Genome Med. 2017;9:1–15. https://doi.org/10.1186/s13073-017-0412-6.
Acknowledgements
This study was conducted when the first author was enrolled in the Genetic Counseling Graduate Program, College of Medicine, University of Cincinnati and Division of Human Genetics, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH. Data extraction from the EMR were provided by the Data & Technology Services team at Cincinnati Children’s Hospital Medical Center. Greg Muthig provided data management support for the project. The business office of the Perinatal Institute at CCHMC provided all charge data. The authors would like to thank Kathleen Collins Ruff, MS, LGC and Ammar Husami, PhD for their support of this work. During the study period, Dan Swarr received funding support from National Institutes of Health K08HL130666.
Author information
Authors and Affiliations
Contributions
LH participated in the design of the study, data acquisition, organization, and interpretation, as well as drafted the initial manuscript and contributed to the final edits of the publication. DK participated in the acquisition, analysis, and interpretation of data as well as provided edits to the final manuscript. KW participated in data collection and organization as well as provided edits to the final manuscript. HH participated in the statistical analysis and interpretation as well as assisted with edits to the manuscript. DTS contributed to the design and implementation of the project, assisted with data interpretation, and critically revised the final manuscript. KS conceptualized and designed the study, coordinated, and supervised data collection as well as analysis, and drafted the manuscript and completed final edits. All authors approved the final manuscript as submitted and agree to be accountable for all aspects of the work.
Corresponding author
Ethics declarations
Competing interests
LH has equity in Exact Sciences Laboratories. KS has participated in a focus group discussion with Illumina. The remaining authors have no conflicts of interest to disclose.
Ethics approval and consent to participate
The need for approval by the CCHMC ethics committee was waived. The study was performed in accordance with the Declaration of Helsinki.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Hagen, L., Khattar, D., Whitehead, K. et al. Detection and impact of genetic disease in a level IV neonatal intensive care unit. J Perinatol 42, 580–588 (2022). https://doi.org/10.1038/s41372-022-01338-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41372-022-01338-0