Abstract
Background
Human norovirus (HuNoV) is a leading cause of disease globally, yet actual incidence is unknown. HuNoV infections are not reportable in the United States, and surveillance is limited to tracking severe illnesses or outbreaks. Wastewater monitoring for HuNoV has been done previously and results indicate it is present in wastewater influent and concentrations are associated with HuNoV infections in the communities contributing to wastewater. However, work has mostly been limited to monthly samples of liquid wastewater at one or a few wastewater treatment plants (WWTPs).
Objective
The objectives of this study are to investigate whether HuNoV GII preferentially adsorbs to wastewater solids, investigate concentrations of HuNoV GII in wastewater solids in wastewater treatment plants across the county, and explore how those relate to clinical measures of disease occurrence. In addition, we aim to develop and apply a mass-balance model that predicts the fraction of individuals shedding HuNoV in their stool based on measured concentrations in wastewater solids.
Methods
We measured HuNoV GII RNA in matched wastewater solids and liquid influent in 7 samples from a WWTP. We also applied the HuNoV GII assay to measure viral RNA in over 6000 wastewater solids samples from 145 WWTPs from across the United States daily to three times per week for up to five months. Measurements were made using digital droplet RT-PCR.
Results
HuNoV GII RNA preferentially adsorbs to wastewater solids where it is present at 1000 times the concentration in influent. Concentrations of HuNoV GII RNA correlate positively with clinical HuNoV positivity rates. Model output of the fraction of individuals shedding HuNoV is variable and uncertain, but consistent with indirect estimates of symptomatic HuNoV infections in the United States.
Impact statement
-
Illness caused by HuNoV is not reportable in the United States so there is limited data on disease occurrence. Wastewater monitoring can provide information about the community spread of HuNoV. Data from wastewater can be available within 24 h of sample receipt at a laboratory. Wastewater is agnostic to whether individuals seek medical care, are symptomatic, and the severity of illness. Knowledge gleaned from wastewater may be used by public health professionals to make recommendations on hand washing, surface disinfection, or other behaviors to reduce transmission of HuNoV, or medical doctors to inform clinical decision making.
Similar content being viewed by others
Introduction
Human norovirus (HuNoV) is a leading cause of gastrointestinal illness globally [1], yet its actual disease burden is largely unknown. This is not only because some infections are asymptomatic, but also because most individuals with symptomatic infections do not have access to, or do not seek clinical care, and those that do are typically not offered diagnostic testing. Testing is generally limited to individuals with severe symptoms or comorbidities. HuNoV is not a reportable disease in many countries, including the United States (US), so the data on the number of confirmed infections is not consistently recorded. Run by the US Center for Disease Control (CDC), Calicinet [2] compiles information about HuNoV outbreaks and genomic sequences, and sentinel laboratories voluntarily submit HuNoV test positivity data to the National Respiratory and Enteric Virus Surveillance System (NREVSS) [3].
Indirect estimates suggest that annually, HuNoV causes between 19 and 21 million symptomatic illnesses and 570–800 deaths in the US [4]. HuNoV infections can be asymptomatic; a study conducted in England found 12% prevalence of asymptomatic HuNoV infections in the population [5] while Teunis et al. [6] report 32% of volunteers infected by HuNoV in a feeding study did not develop symptoms. There are two common genotypes of HuNoV: HuNoV GI and HuNoV GII. HuNoV GII.4 is responsible for the greatest proportion of infections globally [7]. Individuals with inactivated FUT2 enzyme (“non-secretors”) are resistant to infection by some HuNoV genotypes [6, 8].
Wastewater-based epidemiology (WBE) is a low-bias approach for assessing disease burden in a community. Municipal wastewater contains human excretions including feces, urine, saliva, and mucus, that enter the wastewater systems via toilets, showers, sinks, and any other drains in buildings. All individuals using these drains in the sewershed contribute to wastewater. WBE has been used extensively during the COVID-19 pandemic to assess trends in SARS-CoV-2 infections [9], and more recently, it has been used to understand occurrence and trends in Mpox virus [10], influenza A [11, 12], respiratory syncytial virus [13] (RSV), human metapneumovirus, parainfluenza, seasonal coronaviruses, and rhinovirus infections [14]. Concentrations of nucleic-acids from these viruses correlate to clinical measures of disease occurrence, including incident case rates and test positivity rates, in the contributing populations. WBE has also provided insights into Salmonella [15], hepatitis A [16], and poliovirus [17] circulation in communities. Therefore, WBE may be a useful tool for better understanding HuNoV disease occurrence.
HuNoV is shed in high concentrations in stool, where concentrations can be as high as 1011 copies of genomic RNA per gram of stool [18]. Fecal shedding is similar among asymptomatic and symptomatic infections [19]. HuNoV can also be detected in saliva of infected individuals [20] with mean concentrations on the order of 103 copies genomic RNA per ml saliva. Given that HuNoV is found in high concentrations in multiple excretions that contribute to wastewater, it is not surprising that studies have documented HuNoV in wastewater. Eftim et al. [21] carried out a systematic review of HuNoV concentrations in wastewater globally and found that mean concentrations in liquid wastewater influent (globally) are 104.6 copies/liter and tended to be highest in the winter and spring, consistent with the fact that HuNoV infections peak in the winter [22]. Moreover, concentrations of HuNoV GII were higher in wastewater influent than those of HuNoV GI, particularly in North America.
Raw wastewater is a complex mixture of human excretions, food waste, water, trash, and industrial inputs. It contains both a liquid and solid phase. A number of studies suggest that viral nucleic acids and viruses present in raw wastewater tend to adsorb to the solids in wastewater [10, 12, 23]. This has been shown to occur in both experimentally inoculated laboratory studies and in observations of raw wastewater for a wide range of enveloped and non-enveloped viruses [24]. For example, Wolfe et al. [11] showed that influenza A RNA was ~1000 times high in the solids fraction compared to the liquid fraction of raw wastewater while Ye et al. [25] showed in laboratory studies that when added to wastewater, enveloped viruses phi6 and murine hepatitis virus had a higher affinity to wastewater solids than non-enveloped MS2. Da Silva et al. [26] showed that HuNoV RNA was associated with particles in waste stabilization pond influent.Based on this work there is reason to suspect HuNoV may also partition to wastewater solids, but to our knowledge, such data has not been reported in the literature.
Previous work has related HuNoV in wastewater to health. Huang et al. [27] carried out a systematic review and meta-analysis of studies that linked information on HuNoV RNA in wastewater to the health of the contributing populations. They identified seven peer-reviewed studies that reported monthly concentrations of HuNoV RNA in wastewater influent and gastrointestinal infections (measured via a variety of metrics) in the contributing population; an eighth study [28] measured HuNoV RNA weekly in wastewater primary effluent serving approximately 10,000 people and compared their measurements to local gastroenteritis cases. These eight studies generally show good agreement between wastewater influent / primary effluent HuNoV RNA concentrations and measures of gastroenteritis in the communities, suggesting monitoring HuNoV in wastewater may provide useful insights into disease occurrence, and circulating genotypes, in communities. To date, however, no study has examined the occurrence of HuNoV RNA in wastewater solids, and investigated the possibility of using HuNoV RNA in solids for disease surveillance. Additionally, the HuNoV wastewater monitoring studies, to date, have measured HuNoV at a low frequency (primarily monthly). Receiving information on a more frequent basis (daily to weekly) may provide additional insights into disease occurrence and trends in populations.
The goal of the present study is to investigate the use of WBE using wastewater solids to understand HuNoV GII infection occurrence and trends. We focus on HuNoV GII because it has been shown to be more common than HuNoV GI in North America [21]. First, we conducted an experiment to examine the distribution of HuNoV GII RNA between the solid and liquid fractions of wastewater to inform use of the solid fraction for the study. As a first step to investigating the utility of WBE for understand HNoV GII disease occurrence, we first present data on concentrations of HuNoV GII genomic RNA in wastewater solids collected daily for four months at a large wastewater treatment plant (WWTP) that serves 1.5 million people, and compare the data to local clinical HuNoV positivity rates. We then present concentrations of HuNoV GII RNA in wastewater solids collected approximately three times per week from 145 WWTPs located in 26 states for up to 5 months, and compare to national estimates of clinical HuNoV positivity rates. Lastly, we derive a deterministic mass-balance model to estimate the number of individuals shedding HuNoV GII RNA given the concentrations measured in wastewater solids.
Methods
This study was reviewed by the Institutional Review Board (IRB) at Stanford University and the IRB determined that this research does not involve human subjects as defined in 45 CFR 46.102(f) or 21 CFR 50.3(g).
Daily sample collection at San José WWTP
San José (SJ) WWTP (San José-Santa Clara Regional Wastewater Facility) serves 75% (1,500,000 people) of Santa Clara County, California [29]. Daily samples of settled solids were collected between 15 November 2022 and 9 April 2023 (n = 146). Daily samples of influent were collected between 28 February 2023 and 6 March 2023 (n = 7).
Fifty mL of settled solids were collected using sterile technique in clean bottles from the primary clarifier. Twenty-four hour composite samples were collected by combining grab samples every six hours. Samples were stored at 4 °C, transported to the lab, and processed within six hours.
One-hundred mL of twenty-four hour composite influent samples were collected using sterile bottles. Samples were stored at 4 °C, transported to the lab, and then stored for up to 7 days before analysis. Limited degradation of RNA targets over this period of time is expected [30, 31].
Three-day per week sample collection at national WWTPs
Samples were collected typically three times per week at up to 145 WWTPs between November 16, 2022 and April 9, 2023 (Supplementary Fig. S1 and Supplementary Table S1). Samples of settled solids were collected from the primary clarifier, or solids were obtained from raw influent by either using an Imhof cone [32], or allowing the influent to settle for 10–15 min, and using a serological pipette to aspirate the settled solids into a falcon tube. Samples were collected by WWTP staff and sent at 4 °C to our laboratory where they were processed immediately. The time between sample collection and receipt at the lab was typically between 1-3 days, during this time limited degradation of the RNA targets is expected [30, 31]. Supplementary Table S1 provides additional information on the WWTPs including populations served and number and type of samples. In total, these WWTPs serve 10.9% of the US population. Note that the 145 WWTPs include San Jose WWTP. A total of 6911 samples were collected and analyze.
Solid pre-analytical methods
Solids were first dewatered by centrifugation, and then dewatered solids were suspended in DNA/RNA Shield (Zymo Research, Irvine, CA) at a concentration of 0.75 mg (wet weight)/ml. The DNA/RNA shield was spiked with bovine coronavirus (BCoV) vaccine as a RNA recovery control. This concentration of solids was chosen as it was shown to alleviate inhibition in downstream RT-PCR [33]. An additional aliquot of dewatered solids was dried in an oven [29] to determine its dry weight so that measured concentrations of nucleic acid targets could be normalized to gram dry weight. RNA was extracted from 10 or 6 (Supplementary Table S1) replicate aliquots of dewatered settled solids suspended in the DNA/RNA Shield using the Chemagic Viral DNA/RNA 300 kit H96 for the Perkin Elmer Chemagic 360 (Perkin Elmer, Waltham, MA) followed by PCR inhibitor removal with the Zymo OneStep-96 PCR Inhibitor Removal kit (Zymo Research, Irvine, CA) The pre-analytical methods described here are provided in detail in other publications and on protocols.io [29, 33,34,35,36].
Influent pre-analytical methods
For each sample, 10 replicate aliquots were processed using an affinity-based capture method with magnetic hydrogel Nanotrap Particles with Enhancement Reagent 1 (Ceres Nanosciences, Manassas, VA) on 10 mL of sample to concentrate viral particles using a KingFisher Flex system following vendor instructions. RNA was then extracted from the each concentrated aliquot using the MagMAX Viral/Pathogen Nucleic Acid Isolation Kit (Applied Biosystems, Waltham, MA) on the KingFisher Flex platform to obtain purified nucleic acids which were then process through a Zymo OneStep-96 PCR Inhibitor Removal kit (Zymo Research, Irvine, CA).
Digital droplet RT-PCR analytical methods
Each replicate RNA extract from each sample (10 or 6 per sample) was subsequently processed immediately to measure viral RNA concentrations using digital RT-PCR. We quantified the number of copies of the HuNoV GII ORF1-ORF2 junction using a previously established assay that has been shown to be highly specific to HuNoV GII [37]. Ten or 6 replicate wells (Supplementary Table S1) were run for each sample. We also measured concentrations of pepper mild mottle virus (PMMoV) RNA; PMMoV is highly abundant in wastewater globally [38] and is used as an internal recovery and fecal strength control [39]. We also measured concentrations of BCoV RNA. Ten or 2 replicate wells (Supplementary Table S1) were run for each sample for PMMoV and BCoV in duplex). Extraction negative (BCoV spiked buffer, 3 wells) and positive (buffer spiked with positive control cDNA of targets, 1 well) controls, and PCR negative (molecular grade water, 3 wells) and positive controls (gene block cDNA, 1 well) were run on each 96 well plate. Primers and probes are provided in Table 1.
ddRT-PCR was performed on 20 µl samples from a 22 µl reaction volume, prepared using 5.5 µl template, mixed with 5.5 µl of One-Step RT-ddPCR Advanced Kit for Probes (Bio-Rad 1863021), 2.2 µl of 200 U/µl Reverse Transcriptase, 1.1 µl of 300 mM dithiothreitol (DDT) and primers and probes mixtures at a final concentration of 900 nM and 250 nM respectively. Primer and probes for assays were purchased from Integrated DNA Technologies (IDT, San Diego, CA) (Table 1). HuNoV was measured in reactions with undiluted template whereas PMMoV and BCoV assays were run in duplex on template diluted 1:100 in molecular grade water.
Droplets were generated using the AutoDG Automated Droplet Generator (Bio-Rad, Hercules, CA). PCR was performed using Mastercycler Pro (Eppendforf, Enfield, CT) with with the following cycling conditions: reverse transcription at 50 °C for 60 min, enzyme activation at 95 °C for 5 min, 40 cycles of denaturation at 95 °C for 30 s and annealing and extension at 59 °C (for HuNoV GII) or 56 °C (for PMMoV/BCoV) for 30 s, enzyme deactivation at 98 °C for 10 min then an indefinite hold at 4 °C. The ramp rate for temperature changes were set to 2 °C/second and the final hold at 4 °C was performed for a minimum of 30 min to allow the droplets to stabilize. Droplets were analyzed using the QX200 or the QX600 Droplet Reader (Bio-Rad). A well had to have over 10,000 droplets for inclusion in the analysis. All liquid transfers were performed using the Agilent Bravo (Agilent Technologies, Santa Clara, CA).
Thresholding was done using QuantaSoft™ Analysis Pro Software (Bio-Rad, version 1.0.596). In order for a sample to be recorded as positive, it had to have at least 3 positive droplets. Replicate wells were merged for analysis of each sample.
Concentrations of RNA targets were converted to concentrations per dry weight of solids or per volume of influent (copies per gram dry weight (cp/g), or copies per ml (cp/ml), respectively) using dimensional analysis. The total error is reported as standard deviations and includes the errors associated with the Poisson distribution and the variability among the 10 replicates. Three positive droplets across 6 and 10 merged wells corresponds to a concentration between ~300–600 cp/g and ~500–1000 cp/g for solids, respectively, (the range in values is a result of the range in the equivalent mass of dry solids added to the wells) and three positive droplets across 10 merged wells corresponds to 1 cp/ml for influent.
Inhibition testing
Inhibition of PMMoV and BCoV assays is not expected since RNA diluted 1:100 was used as template, and such high dilutions usually alleviate inhibition [40]. However, we tested for inhibition for the HuNoV GII assay in the influent and solids RNA extracts as RNA extracts were run neat as template in those reactions and inhibition could be present. Our previous work with our pre-analytical and analytical workflow suggests no RT-PCR inhibition [33, 41].
RNA extract from an influent sample (collected on 6 March 2023) was selected at random. It was tested for HuNoV GII following exactly the digital droplet RT-PCR analytical methods as follows: RNA extract was run neat as template, and then RNA extract was diluted 1:10 and 1:20 as template. The concentrations of HuNoV (in units of cp/ml) measured in the influent using the different diluted templates were compared to determine whether or not significant inhibition was present.
Two solids samples were chosen at random and rerun according to the preanalytical and analytical methods using the following concentrations of suspension of solids in DNA/RNA shield in the preanalytical processing: 7.5 mg/ml, 15 mg/ml, 37.5 mg/ml, and 75 mg/ml. The concentrations of HuNoV measured in the solids samples (units of cp/g) using the different mass concentration in the suspensions were compared to determine whether or not significant inhibition was present.
HuNoV positivity rate data
There is no data on the incidence and prevalence of HuNoV infections in the United States. As such, we use HuNoV clinical test positivity rates to infer information about HuNoV infections. We used two sources of positivity rate data.
We used positivity rates from Stanford Health Care Clinical Microbiology Laboratory (hereafter “clinical laboratory”) for comparison to the wastewater data from SJ WWTP. The clinical laboratory tests stool specimens of symptomatic patients for HuNoV using the BIOFIRE GI panel (bioMérieux, Inc., Salt Lake City, UT). The clinical laboratory is among the largest in Santa Clara County, CA, where SJ WWTP is located. Although patients may or may not live in the service area of the WWTP, there is a high likelihood they do since the WWTP serves 75% of the county residents. We assumed that the HuNoV positivity rates recorded at the clinical laboratory are reasonable estimates for the positivity rates for residents in the sewershed.
We used national clinical HuNoV positivity rates publicly available through CDC NREVSS [3]. NREVSS compiles positivity rate data from sentinel laboratories across the country and reports positivity rates as 3-week, centered rolling means. Not every state participates in the voluntary reporting program, and not all sentinel laboratories provide HuNoV data to NREVSS; the exact states and laboratories reporting are not publicly available. We used the data as publicly available for the entire US for comparison to the wastewater data from WWTPs included in this study. It is assumed that the positivity rates for HuNoV described in this national data set is a reasonable estimate for the rates in populations served by the WWTPs.
Statistical analyses
Data used in the study are not normally distributed, so we used non-parametric statistics to test hypotheses. We tested the null hypothesis that ratios of HuNoV RNA concentrations and PMMoV RNA in paired solids and liquids are the same using a Wilcoxon signed test. We tested the null hypothesis that the concentration of HuNoV RNA and HuNoV RNA/PMMoV RNA ratio are not associated with HuNoV positivity rates of clinical specimens collected at the clinical laboratory or compiled by CDC NVERSS using Kendall’s tau. Since positivity rates are aggregated by week, we used median wastewater concentrations for the same week as an independent variable. We computed a population-weighted national weighted average of HuNoV and HuNoV/PMMoV across all WWTPs for comparison to the national positivity rates (see SI). We used a p value of 0.05 to assess whether the null hypothesis should be rejected.
Modeling
Previous work established a mass balance model relating the concentration of SARS-CoV-2 RNA in wastewater solids to the number of individuals excreting SARS-CoV-2 RNA into the wastewater [42]. We adopted that model to relate the concentration of HuNoV GII RNA in wastewater solids to the fraction of individuals excreting HuNoV into the wastewater as the mass balance model is not virus specific. The model relies on the assumption that fecal inputs into the wastestream are the main source of viral RNA, and that the fraction of the solids that are fecal in origin can be approximated using the concentration of PMMoV in the solids. According to the model:
where Cn is the concentration of HuNoV RNA in wastewater solids, CPMMoV is the concentration of PMMoV RNA in wastewater solids, CPMMoV_feces is the concentration of PMMoV RNA in feces, kp and Kdp are the PMMoV RNA first-order rate constant and partitioning coefficient, respectively, kn and Kd are the HuNoV RNA first order decay constant and partitioning coefficient, respectively, t is the time sewage spends in the system, including the primary clarifier, prior to sampling, TSS is the total suspended solids in the influent of the treatment plant, Cfeces is the concentration of HuNoV RNA shed in feces, and Fshed is the population fraction shedding HuNoV RNA in feces. The partitioning coefficient describes the ratio of the concentrations of HuNoV RNA in solids and liquids (unit of ml/g).
Given the small decay rate constants reported in the literature for both the HuNoV [30] and PMMoV RNA targets [31, 43], and the short time t (<1 day) that the material spends in the system between toilet and sampling point [42], we neglect the decay term by assuming kn and kp are both small and that kn-kp ~ 0. Equation (1) can then be written:
For the special case where Kd = Kdp, Eq. (2) simplifies to
In applying the model, we assume viral RNA concentrations in settled solids from the primary clarifier are representative of concentrations associated with solids in wastewater, Cn, and that samples represent a temporal composite of inputs.
Arts et al. [44] provide a large dataset characterizing the distribution of CPMMoV_feces. Chan et al. [18] provide data on HuNoV GII shedding in infected individuals within 48 h of symptom onset; we digitized their data (from their Fig. 1A) using plot digitizer (https://plotdigitizer.com/) to characterize the distribution of Cfeces.
We modeled Fshed as a function of Cn/CPMMoV using a Monte Carlo simulation where values for Cfeces and CPMMoV_feces were drawn randomly from the distributions from Chan et al. [18] and Arts et al. [44], respectively. We varied Cn/CPMMoV as log10(Cn/CPMMoV) from −4 to −1 in 0.1 increments. Ten thousand simulations were used to estimate median and interquartile ranges of Fshed for each Cn/CPMMoV.
Results
Quality assurance and control (QA/QC)
Results are reported as suggested in the Environmental Microbiology Minimal Information guidelines [45] (Supplementary Fig. S2). Extraction and PCR negative and positive controls performed as expected (negative and positive, respectively). We observed no evidence of inhibition of the HuNoV GII assay for solids or influent methods (Supplementary Fig. S3).
HuNoV GII and PMMoV RNA in wastewater solids and influent
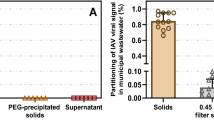
We measured concentrations of indigenous HuNoV GII RNA in matched wastewater influent and solids from SJ WWTP (Fig. 1). Median BCoV recovery in these solids samples was 1.8 (n = 7); values greater than 1 are likely a result of uncertainty in quantifying the amount of BCoV spiked in the DNA/RNA shield. We did not measure BCoV recovery in the influent samples, but according to the manufacturer, recovery of viral RNA from wastewater samples is typically 40–50%.
Matched samples were collected on the same day, but from different locations in the treatment train (influent from the inlet to the WWTP versus solids from the primary clarifier). HuNoV GII RNA concentrations in influent and solids were on the order of 104 cp/ml and 107 cp/g, respectively. The ratio of concentrations in matched solids and influent samples (Kd) varied from 2100 to 3900 ml/g (median = 2500 ml/g, n = 7). Given the enrichment of HuNoV GII in solids relative to influent on an equivalent mass basis, we chose to measure HuNoV GII RNA in solids for our prospective study.
PMMoV RNA concentrations in influent and solids were on the order of 106 cp/ml and 109 cp/g, respectively. The ratio of concentrations in matched solids and influent samples (Kdp) varied from 1600 to 3300 ml/g (median = 2400 ml/g, n = 7). Kdp and Kd were not different (Wilcoxon signed test, p = 0.54).
The ratio of HuNoV GII RNA concentrations and PMMoV RNA concentrations (HuNoV/PMMoV) ranged from 0.026 to 0.029 in influent, and 0.028 to 0.042 in solids. We tested the null hypothesis that HuNoV/PMMoV was not different in influent and solids and the null hypothesis was not rejected (Wilcoxon signed test, p = 0.08).
Prospective daily measurements of HuNoV GII RNA in wastewater solids at SJ WWTP and relation to clinical specimen testing
Median BCoV recovery across the 146 wastewater solids samples for SJ WWTP was 1.7. Recoveries higher than 1 are due to uncertainties associated with quantification of the amount of BCoV spiked into the DNA/RNA shield. No attempt was made to correct measurements by the BCoV recovery owing to complexities and uncertainties associated with measuring virus recovery [46]. Median PMMoV RNA concentrations across samples was 1.5 × 109 cp/g, similar to measurements previously reported for the plant [14]. In addition, PMMoV levels were stable across samples (Supplementary Fig. S4, 25th percentile = 1.3 × 109 cp/g, 75th percentile = 1.8 × 109 cp/g) suggesting consistent fecal strength and RNA extraction efficiency across samples.
HuNoV GII RNA concentrations in wastewater solids varied from 4.5 × 106 to 7.1 × 107 cp/g with median concentrations of 2.3 × 106 cp/g (Fig. 2). Concentrations gradually increased throughout the study with occasional temporary drops in concentration. A similar trend is evident in the ratio of HuNoV GII RNA/ PMMoV RNA.
Top panels: Positivity rate for clinical specimens processed by the clinical laboratory (left) and by the sentinel laboratories participating in NREVSS (right). Middle panels and bottom panel: Concentrations of HuNoV GII RNA and HuNoV GII RNA normalized by PMMoV RNA, respectively. For the SJ WWTP, errors are provided as standard deviations as reported by the analysis software that incorporate variability among 10 replicates and Poisson error; if error bars are not visible, it is because they are smaller than the size of the marker. The error on the ratio is propagated from those of the values in the numerator and denominator. The 5-d trimmed average is shown as well as raw data as a solid line. For the national WWTP, raw data are shown for every WWTP, broken axes are needed to show higher measurements. The population-weighted average lines are shown as solid lines.
The clinical laboratory tested between 62 and 120 (median = 88) specimens per week between 14 November 2022 and 9 April 2023. The weekly HuNoV positivity rate varied from 1% to 16% (median = 7%). The weekly clinical positivity rate was positivity correlated to the weekly median HuNoV GII RNA concentrations in settled solids at the SJ WWTP (Kendall’s tau = 0.47, p = 0.003); the correlation was similar when median weekly HuNoV GII/PMMoV was used for the wastewater variable (Kendall’s tau = 0.45, p = 0.004).
Prospective measurements of HuNoV GII RNA in wastewater solids at 145 WWTPs nationally and relation to clinical specimen testing
Median BCoV recovery across all the samples (n = 8410) was 1.2. Median PMMoV RNA concentrations across samples was 5.4 × 108 (25th percentile = 3 × 108, 75th percentile = 10 × 108 cp/g). HuNoV GII RNA concentrations in wastewater solids varied from 0 to 8.2 × 108 cp/g with median concentrations of 1.5 × 107 cp/g (Fig. 2). The population-weighted average HuNoV GII RNA concentration across all the WWTP (Fig. 2) increased throughout the study period and began to decrease at the end of the time series.
Nationally, weekly-aggregated clinical specimen positivity rates from NREVSS varied from 5% to 17%. The median weekly population weighted average HuNoV GII RNA was positively correlated to the NREVSS positivity rate date (Kendall’s tau = 0.74, p < 10−7). The correlation between HuNoV GII RNA/PMMoV RNA and positivity rate was similar (Kendall’s tau = 0.76, p < 10−7).
Model results
The distributions CPMMoV_feces and Cfeces RNA in units of copies per gram are characterized by log10-means (log10-standard deviations) of 7.72 (1.71) and 8.39 (1.57), respectively. On examination of a Q-Q plot, the distributions are reasonably modeled as log-normal distributions.
The relationship between Cn/CPMMoV and Fshed is linear (Eq. (3), Supplementary Fig. S5). At SJ WWTP, Cn/CPMMoV varied between 0.003 and 0.05 during the study. Based on the model, these minimum and maximum wastewater solids concentrations correspond to Fshed (expressed as percent of individuals shedding) of 0.06% (median model output, 25th percentile = 0.002%, 75th percentile = 2%) and 1% (median model output, 25th percentile = 0.03%, 75th percentile = 35%). The daily estimates of Fshed are displayed in Fig. 3.
The population-weighted average Cn/CPMMoV across all 145 WWTP included in this study ranged from 0.0002 to 0.07. Model output for Fshed, using the population-weighted average Cn/CPMMoV ranges from 0.004% (median model output, 25th percentile = 0.0001%, 75th percentile = 0.2%) to 1.5% (median model output, 25th percentile = 0.04%, 75th percentile = 51%). The daily estimate of Fshed is displayed in Fig. 3.
Discussion
HuNoV GII RNA concentrations in wastewater solids are positively associated with clinical specimen positivity rates both at a single WWTP, and aggregated across WWTPs spanning the entire United States. This finding supports previous work that illustrates HuNoV RNA concentrations in liquid wastewater reflect HuNoV infections in the contributing communities, as summarized in the systematic review by Huang et al. [27]. Whereas most previous studies have studied co-variation in clinical measures of HuNoV infections and wastewater concentrations on monthly scales over the course of a year or more [27], the present work shows the relationship between wastewater solids HuNoV GII RNA concentrations and clinical measures occurrence co-vary over finer and shorter time scales. Wastewater measurements are available as quickly as 24 h after a sample is received at a laboratory, so this work suggests that wastewater testing may provide real-time information on changes in disease occurrence for use by public health officials, clinicians, and the public by arriving in databases before data on clinical infections.
There are limitations associated with the clinical positivity rate data used in this study. It is generated using specimens from patients who may not live in the sewersheds. The specimens collected by the clinical laboratory are from individuals with severe symptomatic illness who sought medical care and the clinicians determined diagnostic testing was needed. Many of these patients likely had comorbidities. The data obtained from CDC NREVSS database are from sentinel laboratories located throughout the US, not necessarily in the states where wastewater data were collected; a list of the precise sentinel laboratories submitting HuNoV data, and the frequency at which they submit data is not publicly available. Therefore, the positivity rates do not necessarily reflect actual community HuNoV infection occurrence in the communities contributed to the wastewater system. Nevertheless, the agreement between the positivity rate and wastewater data lends credence to the wastewater data being reflective of community disease occurrence.
We found HuNoV GII RNA was three orders of magnitude higher in concentration in wastewater solids compared to liquid influent on a mass equivalent basis. We found PMMoV RNA was also concentrated in the solids relative to the liquid influent to a similar extent. This supports previous work that suggests viral nucleic acids partition to the solid phase of wastewater [10,11,12, 23,24,25]. Solids naturally concentrate the viral nucleic acids in the waste stream and can be used as a basis for analysis without the need for time-consuming pre-analytical processes that are not automatable [29]. Because PMMoV RNA and HuNoV GII RNA partition to wastewater solids to similar extents, their ratio in solids and liquid is not different. Additional research is warranted to understand if measurements in wastewater solids and influent can be readily compared after each is normalized to PMMoV concentrations.
We derived a model for relating HuNoV GII RNA concentrations in wastewater solids to the number of individuals in the community shedding HuNoV in feces. Owing to the fact that the partitioning of HuNoV GII and PMMoV RNA was not different, the model simplified significantly resulting in a linear relationship between the fraction of individuals in the sewershed shedding HuNoV GII RNA (Fshed) and the ratio HuNoV GII RNA/PMMoV RNA in solids. HuNoV shedding drops off exponentially with time since day of infection [19], so new infections likely contribute the most to HuNoV GII RNA measured in wastewater, as has also been suspected for SARS-CoV-2 [42]. Therefore, Fshed likely approximates incident HuNoV infections in the contributing population rather than the overall prevalence of infection. Our model used measurements of shedding within the first 48 h after symptom onset from Chan et al. [18] and thus likely represents peak shedding [19]. Individuals shedding higher concentrations of virus in their stool (“super shedders”) will contribute the most to wastewater HuNoV GII RNA.
Summing daily Fshed, estimated from the national data over the duration of the approximate five month study at the median and 75th percentile model outputs suggest that the entire contributing population was infected at least once with HuNoV during the study period. These estimates are probably unrealistic. On the other hand, summing the 25th percentile Fshed over the duration of the five month study suggests 3% of the contribution population was infected with HuNoV GII. This is consistent with indirect estimates that approximately 7% of the US population is infected with symptomatic HuNoV annually [4]. Assuming asymptomatic infections make up 30% of the infections [6], our model at the 25th percentile estimates 2% of the population had a symptomatic infection over 5 months or, scaling linearly, 5% over 12 months. The 25th percentile model output for Fshed is calculated using higher Cn and gives greater weight to “super shedders”. Future efforts are needed to further refine models for estimating incidence and prevalence of disease from wastewater, and fully explore their usefulness. While these results align realistically with other estimates of disease occurrence, the large range in observed peak shedding for HuNoV [19] demonstrates how difficult it may be to model incidence or prevalence of the disease with precision.
WBE can provide information on HuNoV infections in communities and has also been shown to be useful for identifying circulating HuNoV genotypes [47]. It should be noted that WBE should not replace individual testing, as those are used to inform clinical decision making for severely ill patients. However, knowledge gleaned from wastewater may be used by public health professionals to make recommendations on hand washing, surface disinfection, or other behaviors to reduce transmission of HuNoV, or medical doctors to inform clinical decision making. Additional molecular epidemiology applications to HuNoV in wastewater may provide insight into viral genomic diversity and evolution, as well as vaccine development efforts. It should be noted that our study was limited to HuNoV GII, but HuNoV GI likely also contributes to HuNoV disease occurrence.
Data availability
All wastewater data used in this study are available through the Stanford Digital Repository at https://doi.org/10.25740/rk281xb8780.
References
Ahmed SM, Hall AJ, Robinson AE, Verhoef L, Premkumar P, Parashar UD, et al. Global prevalence of norovirus in cases of gastroenteritis: a systematic review and meta-analysis. Lancet Infect Dis. 2014;14:725–30.
United States Center for Disease Control. Reporting and surveillance for norovirus: CaliciNet. 2023. https://www.cdc.gov/norovirus/reporting/calicinet/index.html. Accessed 10 Apr 2023.
United States Center for Disease Control. The National Respiratory and Enteric Virus Surveillance System (NREVSS). 2023. https://www.cdc.gov/surveillance/nrevss/index.html. Accessed 10 Apr 2023.
Hall A, Lopman B, Payne D, Patel M, Gastañaduy P, Vinjé J, et al. Norovirus disease in the United States. Emerg Infect Dis J. 2013;19:1198.
Phillips G, Tam CC, Rodrigues LC, Lopman B. Prevalence and characteristics of asymptomatic norovirus infection in the community in England. Epidemiol Infect. 2010;138:1454–8.
Teunis PFM, Moe CL, Liu P, Miller SE, Lindesmith L, Baric RS, et al. Norwalk virus: how infectious is it? J Med Virol. 2008;80:1468–76.
Matthews JE, Dickey BW, Miller RD, Felzer JR, Dawson BP, Lee AS, et al. The epidemiology of published norovirus outbreaks: a review of risk factors associated with attack rate and genogroup. Epidemiol Infect. 2012;140:1161–72.
Nordgren J, Svensson L. Genetic susceptibility to human norovirus infection: an update. Viruses. 2019;11, https://doi.org/10.3390/v11030226.
Bivins A, North D, Ahmad A, Ahmed W, Alm E, Been F, et al. Wastewater-based epidemiology: global collaborative to maximize contributions in the fight against COVID-19. Environ Sci Technol. 2020;54:7754–7.
Wolfe MK, Yu AT, Duong D, Rane MS, Hughes B, Chan-Herur V. et al. Use of wastewater for Mpox outbreak surveillance in California. N Engl J Med. 2023. https://doi.org/10.1056/NEJMc2213882.
Wolfe MK, Duong D, Bakker KM, Ammerman M, Mortenson L, Hughes B, et al. Wastewater-based detection of two influenza outbreaks. Environ Sci Technol Lett. 2022;9:687–92.
Mercier E, D’Aoust PM, Thakali O, Hegazy N, Jia J-J, Zhang Z, et al. Municipal and neighbourhood level wastewater surveillance and subtyping of an influenza virus outbreak. Sci Rep. 2022;12:15777.
Hughes B, Duong D, White BJ, Wigginton KR, Chan EMG, Wolfe MK, et al. Respiratory Syncytial Virus (RSV) RNA in wastewater settled solids reflects RSV clinical positivity rates. Environ Sci Technol Lett. 2022;9:173–8.
Boehm AB, Hughes B, Duong D, Chan-Herur V, Buchman A, Wolfe MK, et al. Wastewater concentrations of human influenza, metapneumovirus, parainfluenza, respiratory syncytial virus, rhinovirus, and seasonal coronavirus nucleic-acids during the COVID-19 pandemic: a surveillance study. Lancet Microbe. 2023;4:e340–e348.
Diemert S, Yan T. Clinically unreported salmonellosis outbreak detected via comparative genomic analysis of municipal wastewater salmonella isolates. Appl Environ Microbiol. 2019;85:e00139–19.
McCall C, Wu H, O’Brien E, Xagoraraki I. Assessment of enteric viruses during a hepatitis outbreak in Detroit MI using wastewater surveillance and metagenomic analysis. J Appl Microbiol. 2021;131:1539–54.
Manor Y, Handsher R, Halmut T, Neuman M, Bobrov A, Rudich H, et al. Detection of poliovirus circulation by environmental surveillance in the absence of clinical cases in Israel and the Palestinian Authority. J Clin Microbiol. 1999;37:1670.
Chan MCW, Sung JJY, Lam RKY, Chan PKS, Lee NLS, Lai RWM, et al. Fecal viral load and norovirus-associated gastroenteritis. Emerg Infect Dis. 2006;12:1278–80. https://stacks.cdc.gov/view/cdc/16217.
Teunis PFM, Sukhrie FHA, Vennema H, Bogerman J, Beersma MFC, Koopmans MPG. Shedding of norovirus in symptomatic and asymptomatic infections. Epidemiol Infect. 2015;143:1710–7.
Anfruns-Estrada E., Sabrià A., Fuentes C., Sabaté S., Razquin E., Cornejo T. et al. Detection of norovirus in saliva samples from acute gastroenteritis cases and asymptomatic subjects: association with age and higher shedding in stool. Viruses. 2020;12, https://doi.org/10.3390/v12121369.
Eftim SE, Hong T, Soller J, Boehm A, Warren I, Ichida A, et al. Occurrence of norovirus in raw sewage – a systematic literature review and meta-analysis. Water Res. 2017;111:366–74.
Ahmed SM, Lopman BA, Levy K. A systematic review and meta-analysis of the global seasonality of norovirus. PLoS One. 2013;8:e75922.
Kim S, Kennedy LC, Wolfe MK, Criddle CS, Duong DH, Topol A. et al. SARS-CoV-2 RNA is enriched by orders of magnitude in primary settled solids relative to liquid wastewater at publicly owned treatment works. Environ Sci Water Res Technol. 2022, https://doi.org/10.1039/D1EW00826A.
Yin Z, Voice T, Tarabara V, Xagoraraki I. Sorption of human adenovirus to wastewater solids. J Environ Eng. 2018;144:06018008.
Ye Y, Ellenberg RM, Graham KE, Wigginton KR. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ Sci Technol. 2016;50:5077–85.
Da Silva AK, Guyader FSL, Saux J-CL, Pommepuy M, Montgomery MA, Elimelech M. Norovirus removal and particle association in a waste stabilization pond. Environ Sci Technol. 2008;42:9151–7.
Huang Y, Zhou N, Zhang S, Yi Y, Han Y, Liu M, et al. Norovirus detection in wastewater and its correlation with human gastroenteritis: a systematic review and meta-analysis. Environ Sci Pollut Res. 2022;29:22829–42.
Kazama S, Miura T, Masago Y, Konta Y, Tohma K, Manaka T. et al. Environmental surveillance of norovirus genogroups I and II for sensitive detection of epidemic variants. Appl Environ Microbiol. 2017. https://doi.org/10.1128/AEM.03406-16.
Wolfe MK, Topol A, Knudson A, Simpson A, White B, Duc V, et al. High-frequency, high-throughput quantification of SARS-CoV-2 RNA in wastewater settled solids at eight publicly owned treatment works in Northern California shows strong association with COVID-19 Incidence. mSystems. 2021;6:e00829–21.
Guo Y, Li J, O’Brien J, Sivakumar M, Jiang G. Back-estimation of norovirus infections through wastewater-based epidemiology: a systematic review and parameter sensitivity. Water Res. 2022;219:118610.
Burnet J-B, Cauchie H-M, Walczak C, Goeders N, Ogorzaly L. Persistence of endogenous RNA biomarkers of SARS-CoV-2 and PMMoV in raw wastewater: Impact of temperature and implications for wastewater-based epidemiology. Sci Total Environ. 2023;857:159401.
AWWA. Standard methods for the examination of water and wastewater. 21st ed. Baltimore: American Public Health Association, American Water Works Association, Water Environment Federation; 2005.
Huisman JS, Scire J, Caduff L, Fernandez-Cassi X, Ganesanandamoorthy P, Kull A, et al. Wastewater-based estimation of the effective reproductive number of SARS-CoV-2. Environ Health Perspect. 2022;130:057011–1.
Topol A, Wolfe M, White B, Wigginton K, Boehm A. High throughput SARS-COV-2, PMMOV, and BCoV quantification in settled solids using digital RT-PCR. protocols.io, 2021. https://doi.org/10.17504/protocols.io.btywnpxe.
Topol A, Wolfe M, White B, Wigginton K, Boehm A. High Throughput pre-analytical processing of wastewater settled solids for SARS-CoV-2 RNA analyses. protocols.io, 2021. https://doi.org/10.17504/protocols.io.btyqnpvw.
Topol A, Wolfe M, Wigginton K, White B, Boehm A. High Throughput RNA Extraction and PCR Inhibitor Removal of Settled Solids for Wastewater Surveillance of SARS-CoV-2 RNA. protocols.io, 2021. https://doi.org/10.17504/protocols.io.btyrnpv6.
Loisy F, Atmar RL, Guillon P, Le Cann P, Pommepuy M, Le Guyader F. Real-time RT-PCR for norovirus screening in shellfish. J Virological Methods. 2005;123:1–7.
Symonds EM, Nguyen KH, Harwood VJ, Breitbart M. Pepper mild mottle virus: a plant pathogen with a greater purpose in (waste)water treatment development and public health management. Water Res. 2018;144:1–12.
McClary-Gutierrez JS, Aanderud ZT, Al-faliti M, Duvallet C, Gonzalez R, Guzman J, et al. Standardizing data reporting in the research community to enhance the utility of open data for SARS-CoV-2 wastewater surveillance. Environ Sci Water Res Technol. 2021;7:1545–51.
Cao Y, Griffith JF, Dorevitch S, Weisberg SB. Effectiveness of qPCR permutations, internal controls and dilution as means for minimizing the impact of inhibition while measuring Enterococcus in environmental waters. J Appl Microbiol. 2012;113:66–75.
Boehm AB, Wolfe MK, Wigginton KR, Bidwell A, White BJ, Hughes B, et al. Human viral nucleic acids concentrations in wastewater solids from Central and Coastal California USA. Sci Data. 2023;10:396.
Wolfe MK, Archana A, Catoe D, Coffman MM, Dorevich S, Graham KE, et al. Scaling of SARS-CoV-2 RNA in settled solids from multiple wastewater treatment plants to compare incidence rates of laboratory-confirmed COVID-19 in their sewersheds. Environ Sci Technol Lett. 2021. https://doi.org/10.1021/acs.estlett.1c00184.
Roldan-Hernandez L, Graham KE, Duong D, Boehm AB. Persistence of endogenous SARS-CoV-2 and pepper mild mottle virus RNA in wastewater-settled solids. ACS EST Water. 2022. https://doi.org/10.1021/acsestwater.2c00003.
Arts PJ, Kelly JD, Midgley CM, Anglin K, Lu S, Abedi GR, et al. Longitudinal and quantitative fecal shedding dynamics of SARS-CoV-2, pepper mild mottle virus, and crAssphage. mSphere. 2023;0:e00132–23.
Borchardt MA, Boehm AB, Salit M, Spencer SK, Wigginton KR, Noble RT. The Environmental Microbiology Minimum Information (EMMI) guidelines: qPCR and dPCR quality and reporting for environmental microbiology. Environ Sci Technol. 2021;55:10210–23.
Kantor RS, Nelson KL, Greenwald HD, Kennedy LC. Challenges in measuring the recovery of SARS-CoV-2 from wastewater. Environ Sci Technol. 2021;55:3514–9.
Lee C-C, Chiu C-H, Lee H-Y, Tsai C-N, Chen C-L, Chen S-Y. Clinical and virological characteristics of viral shedding in children with norovirus gastroenteritis. J Microbiol Immunol Infect. 2022;55:1188–94.
Acknowledgements
We acknowledge the numerous people across the country who contributed to wastewater sample collection, Payal Sakar at SJ WWTP for her support and contribution to the work, and Allegra Koch for her support with the literature review. This research was performed on the ancestral and unceded lands of the Muwekma Ohlone people. We pay our respects to them and their Elders, past and present, and are grateful for the opportunity to live and work here.
Funding
This work is supported by gifts from the CDC Foundation and the Sergey Brin Family Foundation.
Author information
Authors and Affiliations
Contributions
ABB: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Resources, Data Curation, Writing - Original Draft, Writing - Review & Editing, Visualization, Supervision, Project administration, Funding acquisition. MKW: Conceptualization, Methodology, Data Curation, Validation, Writing - Review & Editing. BH: Methodology, Validation, Investigation, Data Curation, Writing - Review & Editing. DD: Methodology, Validation, Investigation, Data Curation, Writing - Review & Editing. AB: Data Curation, Writing - Original Draft, Writing - Review & Editing, Visualization. BW: Methodology, Validation, Investigation, Resources, Data Curation, Writing - Review & Editing, Supervision. NB: Data Curation, Writing - Review & Editing.
Corresponding author
Ethics declarations
Competing interests
Bradley White, Dorothea Duong, and Bridgette Hughes are employees of Verily Life Sciences. The other authors have no competing interests.
Ethical approval
This study was reviewed by the Institutional Review Board (IRB) at Stanford University and the IRB determined that this research does not involve human subjects as defined in 45 CFR 46.102(f) or 21 CFR 50.3(g).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Boehm, A.B., Wolfe, M.K., White, B.J. et al. Human norovirus (HuNoV) GII RNA in wastewater solids at 145 United States wastewater treatment plants: comparison to positivity rates of clinical specimens and modeled estimates of HuNoV GII shedders. J Expo Sci Environ Epidemiol (2023). https://doi.org/10.1038/s41370-023-00592-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41370-023-00592-4