Abstract
Background/objective:
N6-methyladenosine (m6A) modification of mRNA plays an important role in regulating adipogenesis. However, its underlying mechanism remains largely unknown.
Subjects/methods:
Using Jinhua and Landrace pigs as fat and lean models, we presented a comprehensive transcriptome-wide m6A profiling in adipose tissues from these two pig breeds. Two differentially methylated genes were selected to explore the mechanisms of m6A-mediated regulation of gene function.
Results:
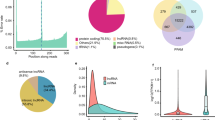
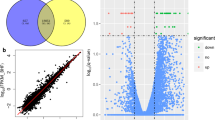
The ratio of m6A/A in the layer of backfat (LB) was significantly higher in Landrace than that in Jinhua. Transcriptome-wide m6A profiling revealed that m6A modification on mRNA occurs in the conserved sequence motif of RRACH and that the pig transcriptome contains 0.53–0.91 peak per actively expressed transcript. The relative density of m6A peaks in the 3′UTR were higher than in 5′UTR. Genes with common m6A peaks from both Landrace (L-LB) and Jinhua (J-LB) were enriched in RNA splicing and cellular lipid metabolic process. The unique m6A peak genes (UMGs) from L-LB were mainly enriched in the extracellular matrix (ECM) and collagen catabolic process, whereas the UMGs from J-LB are mainly involved in RNA splicing, etc. Lipid metabolism processes were not significantly enriched in the UMGs from L-LB or J-LB. Uncoupling protein-2 (UCP2) and patatin-like phospholipase domain containing 2 (PNPLA2) were two of the UMGs in L-LB. Synonymous mutations (MUT) were conducted to reduce m6A level of UCP2 and PNPLA2 mRNAs. Adipogenesis test showed that UCP2-MUT further inhibited adipogenesis, while PNPLA2-MUT promoted lipid accumulation compared with UCP2-WT and PNPLA2-WT, respectively. Further study showed m6A negatively mediates UCP2 protein expression and positively mediates PNPLA2 protein expression. m6A modification affects the translation of PNPLA2 most likely through YTHDF1, whereas UCP2 is likely neither the target of YTHDF2 nor the target of YTHDF1.
Conclusion:
Our data demonstrated a conserved and yet dynamically regulated m6A methylome in pig transcriptomes and provided an important resource for studying the function of m6A epitranscriptomic modification in obesity development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat Rev Genet. 2014;15:293–306.
Wu R, Jiang D, Wang Y, Wang X. N (6)-methyladenosine (m(6)A) methylation in mRNA with a dynamic and reversible epigenetic modification. Mol Biotechnol. 2016;58:450–459.
Krug RM, Morgan MA, Shatkin AJ. Influenza viral mRNA contains internal N6-methyladenosine and 5’-terminal 7-methylguanosine in cap structures. J Virol. 1976;20:45–53.
Bodi Z, Button JD, Grierson D, Fray RG. Yeast targets for mRNA methylation. Nucleic Acids Res. 2010;38:5327–5335.
Dominissini D, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206.
Bokar JA. The biosynthesis and functional roles of methylated nucleosides in eukaryotic mRNA. In: Grosjean H, editor. Fine-tuning of RNA functions by modification and editing. Berlin, Heidelberg: Springer; 2005. p. 141–177..
Harper JE, Miceli SM, Roberts RJ, Manley JL. Sequence specificity of the human mRNA N6-adenosine methylase in vitro. Nucleic Acids Res. 1990;18:5735–5741.
Meyer KD, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646.
Liu J, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93–95.
Wang X, Zhu L, Chen J, Wang Y. mRNA m(6)A methylation downregulates adipogenesis in porcine adipocytes. Biochem Biophys Res Commun. 2015;459:201–207.
Zhao X, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014;24:1403–1419.
Schook L, et al. Swine in biomedical research: creating the building blocks of animal models. Anim Biotechnol. 2005;16:183–190.
Vodicka P, et al. The miniature pig as an animal model in biomedical research. Ann NY Acad Sci. 2005;1049:161–171.
Ibrahim Z, et al. Selected physiologic compatibilities and incompatibilities between human and porcine organ systems. Xenotransplantation. 2006;13:488–499.
Turk JR, Henderson KK, Vanvickle GD, Watkins J, Laughlin MH. Arterial endothelial function in a porcine model of early stage atherosclerotic vascular disease. Int J Exp Pathol. 2005;86:335–345.
Turk JR, Laughlin MH. Physical activity and atherosclerosis: which animal model? Can J Appl Physiol. 2004;29:657–683.
Jia GF, et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–887.
Dominissini D, Moshitch-Moshkovitz S, Salmon-Divon M, Amariglio N, Rechavi G. Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nat Protoc. 2013;8:176–189.
Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589.
Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–U174.
He S, et al. mRNA N6-methyladenosine methylation of postnatal liver development in pig. PLoS One. 2017;12:e0173421.
Luo GZ, et al. Unique features of the m(6)A methylome in Arabidopsis thaliana. Nat Commun. 2014;5:5630.
Perry RP, Kelley DE, Friderici K, Rottman F. The methylated constituents of L cell messenger RNA: evidence for an unusual cluster at the 5’ terminus. Cell. 1975;4:387–394.
Wan Y, et al. Transcriptome-wide high-throughput deep m(6)A-seq reveals unique differential m(6)A methylation patterns between three organs in Arabidopsis thaliana. Genome Biol. 2015;16:272.
Hausman GJ. Meat Science and Muscle Biology Symposium: the influence of extracellular matrix on intramuscular and extramuscular adipogenesis. J Anim Sci. 2012;90:942–949.
Carroll SM, Narayan P, Rottman FM. N6-methyladenosine residues in an intron-specific region of prolactin pre-mRNA. Mol Cell Biol. 1990;10:4456–4465.
Shimba S, Bokar JA, Rottman F, Reddy R. Accurate and efficient N-6-adenosine methylation in spliceosomal U6 small nuclear RNA by HeLa cell extract in vitro. Nucleic Acids Res. 1995;23:2421–2426.
Xiao W, et al. Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol Cell. 2016;61:507–519.
Zhao X, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014;24:1403–1419.
Nagy TR, Blaylock ML, Garvey WT. Role of UCP2 and UCP3 in nutrition and obesity. Nutrition. 2004;20:139–144.
Kraemer FB, Shen WJ. Hormone-sensitive lipase: control of intracellular tri-(di-)acylglycerol and cholesteryl ester hydrolysis. J Lipid Res. 2002;43:1585–1594.
Zimmermann R, et al. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 2004;306:1383–1386.
Shi H, et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017;27:315–328.
Li A, et al. Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res. 2017;27:444–447.
Wang X, et al. N-6-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399.
Wang X, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120.
Li Y, et al. Transcriptome-wide N(6)-methyladenosine profiling of rice callus and leaf reveals the presence of tissue-specific competitors involved in selective mRNA modification. RNA Biol. 2014;11:1180–1188.
Toda C, Diano S. Mitochondrial UCP2 in the central regulation of metabolism. Best Pract Res Clin Endocrinol Metab. 2014;28:757–764.
Jia JJ, Zhang X, Ge CR, Jois M. The polymorphisms of UCP2 and UCP3 genes associated with fat metabolism, obesity and diabetes. Obes Rev. 2009;10:519–526.
Nadler ST, et al. The expression of adipogenic genes is decreased in obesity and diabetes mellitus. Proc Natl Acad Sci Usa. 2000;97:11371–11376.
Donadelli M, Dando I, Fiorini C, Palmieri M. UCP2, a mitochondrial protein regulated at multiple levels. Cell Mol Life Sci. 2014;71:1171–1190.
Pheiffer C, et al. Expression of UCP2 in Wistar rats varies according to age and the severity of obesity. J Physiol Biochem. 2016;72:25–32.
Azzu V, Brand MD. The on-off switches of the mitochondrial uncoupling proteins. Trends Biochem Sci. 2010;35:298–307.
Rousset S, et al. UCP2 is a mitochondrial transporter with an unusual very short half-life. FEBS Lett. 2007;581:479–482.
Ren Y, et al. Breed difference of porcine Sirtuin 1, adipose triglyceride lipase (ATGL) and hormone sensitive lipase (HSL). Livest Sci. 2013;158:199–205.
Acknowledgements
This work is supported by the National Natural Science Foundation of China (Grant No. 31572413) and the Special Fund for Cultivation and Breeding of New Transgenic Organism (No. 2014ZX0800949B).
Author contributions
YW and XW conceived the project; YW and XW designed most experiments; BS performed data analyses; XW, QJ, RW, and MC performed the experiment; XW, QJ, and HS wrote the paper with suggestions from YW.
Accession codes
The high-throughput data used in this study are deposited in the NCBI GEO database with accession number GSE87625.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
These authors contributed equally: Xinxia Wang, Baofa Sun, Qin Jiang, Ruifan Wu.
Rights and permissions
About this article
Cite this article
Wang, X., Sun, B., Jiang, Q. et al. mRNA m6A plays opposite role in regulating UCP2 and PNPLA2 protein expression in adipocytes. Int J Obes 42, 1912–1924 (2018). https://doi.org/10.1038/s41366-018-0027-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41366-018-0027-z
This article is cited by
-
Profiling of N6-methyladenosine methylation in porcine longissimus dorsi muscle and unravelling the hub gene ADIPOQ promotes adipogenesis in an m6A-YTHDF1–dependent manner
Journal of Animal Science and Biotechnology (2023)
-
Insights into N6-methyladenosine and programmed cell death in cancer
Molecular Cancer (2022)
-
mRNA m5C inhibits adipogenesis and promotes myogenesis by respectively facilitating YBX2 and SMO mRNA export in ALYREF-m5C manner
Cellular and Molecular Life Sciences (2022)
-
Current insights into the implications of m6A RNA methylation and autophagy interaction in human diseases
Cell & Bioscience (2021)
-
N6-Adenosine Methylation (m6A) RNA Modification: an Emerging Role in Cardiovascular Diseases
Journal of Cardiovascular Translational Research (2021)