Abstract
Ankyrin proteins act as molecular scaffolds and play an essential role in regulating cellular functions. Recent evidence has implicated the ANK3 gene, encoding ankyrin-G, in bipolar disorder (BD), schizophrenia (SZ), and autism spectrum disorder (ASD). Within neurons, ankyrin-G plays an important role in localizing proteins to the axon initial segment and nodes of Ranvier or to the dendritic shaft and spines. In this review, we describe the expression patterns of ankyrin-G isoforms, which vary according to the stage of brain development, and consider their functional differences. Furthermore, we discuss how posttranslational modifications of ankyrin-G affect its protein expression, interactions, and subcellular localization. Understanding these mechanisms leads us to elucidate potential pathways of pathogenesis in neurodevelopmental and psychiatric disorders, including BD, SZ, and ASD, which are caused by rare pathogenic mutations or changes in the expression levels of ankyrin-G in the brain.
Similar content being viewed by others
Introduction
Ankyrin-repeat (ANK) proteins perform various biological functions and are phylogenetically conserved. They are involved in many cellular processes, such as intercellular connections, signal transduction, cell cycle regulation, vesicle transport, inflammatory responses, cytoskeletal integrity, and transcriptional regulation1. In humans, 270 ANK proteins have been reported and are significantly correlated with psychiatric risk factors such as BD, ASD, and SZ2. An enrichment of ankyrin-repeat domain (ANKRD)-containing protein-encoding genes in BD genome-wide association studies (GWAS) and de novo variants related to ASDs and SZ indicates their broader role in neuropsychiatric diseases3,4,5,6,7.
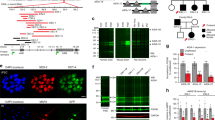
We reanalyzed the previously reported correlation between ANKRD-containing proteins and psychiatric risk genes2 after adding several recently reported BD risk genes7 and 1003 of the ASD risk genes described by the Simons Foundation Autism Research Initiative (SFARI). Significant enrichment of ANKRD-containing proteins was corroborated in ASD and SZ (Fig. 1a). The ANKRD family includes prominent synaptic proteins such as ankyrin-R, -G, and -B (encoded by ANK1/2/3), as well as SHANK1/2/3 and TRANK1, which are associated with BDs, ASDs, and SZ. Moreover, ankyrin-G and SHANK1/2 were confirmed to show comorbidities with ASDs in BDs or SZ. Human ANK3 is related to several neuropsychiatric disorders, including BD, ASD, intellectual disability (ID), and attention-deficit/hyperactivity disorder8,9,10,11. According to the protein–protein interaction (PPI) network of ankyrin-G from the STRING database, 56 out of 200 related genes (12 genes in SZ, 12 genes in BD, 45 genes in ASD) were identified as significantly included in each psychiatric disorder. Among these, 13 factors were confirmed to exhibit comorbidity (Fig. 1b, c).
a Cytoscape analysis of the protein–protein interaction (PPI) network of psychiatric risk genes from GWAS and SFARI and ANKRD-containing proteins retrieved from UniProt and STRING. The large circular nodes indicate psychiatric risk genes, and edge thickness represents the connection score of the connected proteins (0.400 < score < 1.000). *p < 0.05; hypergeometry test (edited from Yoon et al.2). b Enrichment of SZ risk factors (n = 395) (top), BD risk factors (n = 155) (bottom) identified through GWAS and ASD risk factors (n = 1003) (center) from SFARI among the PPI network of ankyrin-G from STRING (n = 200). ***p < 0.001; hypergeometry test. c The representative majority of a subnetwork is visualized and annotated by Cytoscape. The color-coded nodes indicate the score factor (0.100 < score < 1.000). SFARI Simons Foundation Autism Research Initiative; STRING Search Tool for the Retrieval of Interacting Genes/Proteins.
In this review, we explore the genetics, spatiotemporal expression patterns, PPI networks, and pathophysiology related to ankyrin-G, which has been highlighted as a high-confidence psychiatric risk gene in extensive genomic studies. Furthermore, we will discuss the possibility of repurposing existing drugs and using current knowledge of the posttranslational modifications of ankyrin-G to develop new drug candidates to treat psychiatric disorders.
Genetics of ANK3 and neuropsychiatric disorders
Bipolar disorder
In 2008, two independent GWASs highlighted single nucleotide polymorphisms (SNPs) in ANK3, including rs10994336, rs1938526, and rs9804190, as risk factors for BD8,12. A weak linkage disequilibrium between the SNPs was identified, indicating possibly independent contributions of rs10994336 and rs9804190 to the risk of BD9. The association between ANK3 and BD has since been replicated in multiple follow-up GWASs, identifying a range of additional SNPs to further confirm the link between ankyrin-G and psychiatric disease (Supplementary Table 1). The most recent large-scale study analyzed 41,917 individuals with BD and 371,549 controls and again confirmed ANK3 as a leading BD risk locus (odds ratio = 1.125, p = 1.1 × 10−11). This study found a large range of SNPs spanning ANK3 to be associated with BD, all in linkage disequilibrium with rs10994415, the most significant SNP7. In addition to the strong genetic links, ankyrin-G polymorphisms have been linked to certain cognitive and anatomic BD subphenotypes; allelic variation in ANK3 was associated with deficits in sustained attention13,14, decision-making15, working memory16, and verbal comprehension and logical memory17. Moreover, magnetic resonance imaging (MRI) studies showed associations of rs10994336, rs9804190, and rs10761482 with the hyperactivation or hypoactivation of specific brain areas16,18 and atrophy of white and gray matter in defined brain structures15,19,20.
Despite the strong genetic links between ANK3 and BD, the mechanisms by which risk SNPs affect the expression/function of ankyrin-G and cause disease are not well understood. However, studies have shown that risk SNPs in ANK3 are correlated with a change in ANK3 mRNA levels in the blood21 and decreased expression of a specific mRNA isoform containing exon 1B in the cerebellum22 and that the rs10994336 SNP is associated with a lower methylation level at the CpG site cg0217218223. Moreover, a minor isoform of ANK3 containing exon ENSE00001786716 is increased in BD patients24, whereas the loss-of-function mutation rs41283526, which disables correct splicing, has a protective effect25. Interestingly, miR34a and miR10b-5p, two microRNAs targeting ANK3 mRNA, were overexpressed in the postmortem brain26 and iPSCs27 in BD patients. Altogether, these data suggest a critical role of ANK3 isoform expression as a risk factor in BD.
Rare variants in the ANK3 gene, including missense mutations in the promotor or the 3’ untranslated region, have been identified in BD (Supplementary Table 1) from whole-genome or whole-exome sequencing studies28,29. Intriguingly, the effect of the Trp1989Arg mutation, found in a family of BD patients, involves disruption of the ankyrin-G and GABARAP interaction, resulting in a decrease in GABAergic synapses30.
Schizophrenia
A GWAS from 2010 found that SNP rs10761482 in the ANK3 gene was nominally associated with SZ. However, it did not surpass the threshold for genome-wide significance19,31,32. Because SZ and BD share risk alleles33, several studies have investigated SNP associations from previous BD GWASs in SZ patient cohorts34,35,36,37 (Supplementary Table 1). Multiple studies have reported that SNPs in the ANK3 locus are linked to SZ endophenotypes, such as deficits in working memory38,39, proneness to anhedonia40, and cognitive impairment associated with a reduction in gray matter41. The most recent large-scale GWAS for SZ did not show a significant association between ANK3 and SZ42. However, a recent GWAS from patients with psychotic experiences, including those with BD and SZ, found an association for two ANK3 loci, an intronic variant (rs10994278) and an intergenic variant (rs549656827)43. Thus, the link between SZ and ANK3 remains unclear, and further work is required to determine whether a link between ANK3 and SZ or SZ subtypes exists.
Autism spectrum disorders
In contrast with BD and SZ, most genetic studies on ASD have focused on rare de novo mutations rather than GWAS; therefore, no common variants have yet been found linking ANK3 with ASD, but a multitude of rare de novo mutations in ASD probands have been reported (Supplementary Table 1). Missense mutations were found in exons 5, 6, 30, and 37 of ANK3, and several frameshift mutations were also identified4,10,11,44,45,46,47,48,49,50,51. In addition, rare mutations in ANK3, CREBBP, and SEMA6B were identified in an ASD patient, inherited from both parents. Each mutation was associated with increased gene expression, and the mutations could act synergistically to modulate disease severity45.
Multiple isoforms of ankyrin-G in the brain
The ANK3 gene is localized to human chromosome 10q21 between 61.4 and 62.2 million base pairs. Five alternative first exons have been identified for human ANK3, namely, exons 1a, 1b, 1e, 1f, and 1s22. Isoforms including exon 1e and exon 1b were found to be expressed at similar levels in the frontal cortex and cingulate cortex and accounted for almost all ANK3 expression. However, ANK3 exon 1b was expressed at approximately 3−4-fold higher levels in the cerebellum than in the frontal or cingulate cortex22. In C57BL/6J mice, exon 1b expression was also highest in the cerebellum and was substantial in the dentate gyrus and all other regions examined (hippocampus, amygdala, orbitofrontal cortex, PFC, and striatum)52. Ank3 exon 1b transcripts were expressed at higher levels in all brain regions than the alternative leading exon 1e, expressed in transcripts found in the brain and other tissues53. Moreover, exon 1b expression was limited to oligodendrocytes, whereas exon 1b- and 1e-containing ankyrin-Gs were expressed in neurons54.
There are five isoforms for human ANK3 in the RefSeq database encoding ankyrin-G. Three of these, one weighing 202 kDa and two weighing 204 kDa, have a domain structure that includes ANK repeat, spectrin-binding, and regulatory domains. These three protein isoforms are collectively referred to as ankyrin-G 190 because they migrate at 190 kDa by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE). Ankyrin-G 190 localizes to postsynaptic sites and dendritic spines, modulating dendritic spine morphology and synaptic transmission at glutamatergic synapses55. The fourth form has an additional large exon 37 (Ensembl ID ENSE00000997921), which encodes a serine-rich domain. This large exon can be spliced in 2 distinct ways, and the resulting transcripts are referred to as the ankyrin-G 270/480 isoforms (Fig. 2a). The isoform with the shorter ANK3 exon 37 (ankyrin-G 270) is preferentially expressed in the frontal and cingulate cortex, whereas the isoform with the longer ANK3 exon 37 (ankyrin-G 480) has higher expression in the cerebellum22. Alternative splicing modifications of exon 37 are detected only in brain tissue, and these different isoforms are known to have diverse functions at different cellular locations. The fifth isoform has the lowest molecular weight, 110 kDa, and completely lacks the ankyrin-repeat domain. The expression of this isoform, starting from exon 1 s, within specific regions in the brain is poorly understood. Functionally, it has been previously reported that this isoform regulates endocytosis and lysosomal degradation56. Of note, exon 41 (Ensembl ID ENSE00001765562) has been identified as an exon inserted into the 190 and 110 kDa isoforms of ankyrin-G, and the Homer1b/c or p85 subunit of PI3K proteins are known to bind to this inserted site56,57.
a A diagram of ANK3 transcription start sites and alternative splicing. b Expression patterns of the ANK3 gene in the human brain (adapted from The Human Protein Atlas). c A schematic summary of the trajectories of the different ankyrin-G isoforms in the mouse brain. Expression levels were measured as log10 of western blot intensities from Yoshimura et al.61.
Expression patterns of ankyrin-G in the brain
ANK3 is most highly expressed in the cerebellum but is generally expressed in other brain regions, such as the cerebral cortex, hippocampus, corpus callosum, and hypothalamus (Fig. 2b; from The Human Protein Atlas project). These results are consistent with previous reports analyzing the expression pattern of ANK3 mRNA in the brains of mice52,53. In 3-month-old wild-type mice, ankyrin-G is expressed in 40% of cells in the somatosensory cortex from layer II to V and in 57% of cells in the CA3 of the hippocampus2. Analyses of conditional Ank3 knockout mice (AnkG cKO) with homozygous loss of ankyrin-G in forebrain pyramidal neurons, 70% in the somatosensory cortex and 95% in the CA3 of the hippocampus, indicate that ankyrin-G-expressing cells could be excitatory neurons58,59. These decreases were also observed in the piriform and cingulate cortices. A certain percentage of cells positive for ankyrin-G coexpressed with parvalbumin was observed in the AnkG cKO brain, confirming that ankyrin-G is also expressed in inhibitory neurons58. Considering the critical role of synaptic dysfunction in neuropsychiatric disorders, we suggest that at least some phenotypes arise from ankyrin-G-mediated disorders, alterations in the excitatory−inhibitory (E/I) balance, and circuit disturbances in forebrain regions. In addition, specific ankyrin-G in oligodendrocytes is essential for the enrichment of myelin sheaths surrounding the nodes of Ranvier (NoR)54.
Developmental expression patterns of ankyrin-G isoforms
The developmental transcriptome dataset from BrainSpan illuminates how the expression of exons changes from infancy to adulthood. Human exon 25 (Ensembl ID ENSE00001786716) from the RNA-seq dataset has a low level of expression during infancy. However, its expression increases at the age of 11 and reaches a plateau throughout adolescence and adulthood. The expression of exon 24 (Ensembl ID ENSE00000997958) is also low, similar to that of exon 25, in the fetal period. However, its expression gradually increases from childhood to reach a considerably higher level throughout adolescence and adulthood25.
Immunohistochemical observation of the PFC of a rhesus monkey showed that ankyrin-G expression in the axon initial segment (AIS) decreased from birth until 3 months of age. The observation then shows a pattern of stabilizing expression from adolescence to adulthood60. These results were consistent with the results of a western blot observations of mouse whole-brain lysate. Ankyrin-G 480, expressed in the AIS, continuously decreases after birth and stabilizes through adolescence, whereas ankyrin-G 270 is evenly expressed throughout life61. Interestingly, in the case of ankyrin-G 190, the expression continuously increases after birth, and the expression level is maintained at a constant level after adolescence and into adulthood (Fig. 2c). These results suggest that three major isoforms in the brain, ankyrin-G 190 and ankyrin-270/480, play their respective roles at distinct locations in neurons throughout the developmental period.
Structure of ankyrin-G and its binding partners
ANKRD
Ankyrin-G, as a scaffolding protein, plays a pivotal role in regulating the localization of ion channels, membrane transporters, cell adhesion molecules, and cytoskeletal proteins. Here, we introduce the ANKRD-binding partners of ankyrin-G.
Ion channels/transporters/pumps
It has been reported that ankyrin-G plays an important role in response to physiological signals by interacting with various types of membrane transporters through ANKRD. Cyclic nucleotide-gated (CNG) channels, such as the rod photoreceptor cGMP-gated cation channel, can localize to the plasma membrane by binding to ankyrin-G through the cytoplasmic C-terminal domain of the channel β1 subunit62. Ankyrin-G is exclusively localized to rod outer segments (ROSs) and is required for the transport of CNG-β1 and ROS morphogenesis. Ankyrin-G also modulates the localization of other membrane transporters, such as sodium/potassium-transporting ATPase subunit alpha-1 (Na +/K + ATPase α1), anion exchanger 1 (AE1), and ammonium transporter Rh type B (RhBG), via interaction with their cytosolic domains63,64,65,66. Exons 13–24 of ANKRD are interaction sites for AE1 and RhBG66. Binding to ankyrin-G is necessary for voltage-gated sodium channels to form clusters at the AIS and the NoR. Ankyrin-G uses its ANKRD to bind to members of the Nav1 family by recognizing a 9-amino-acid motif ((V/A)P(I/L)AXXE(S/D)D) in the cytosolic loop that links transmembrane domains II and III in these channels67,68. Ankyrin-G additionally interacts with potassium channels Kv7.2 (KCNQ2) and Kv7.3 (KCNQ3) via a conserved stretch of 10 amino acids, which is similar to the ankyrin-G-binding sequence in Nav1 channels69.
Cell adhesion molecules
Ankyrin-G is known to regulate the organization of the AIS and NoR through interaction with cell adhesion molecules such as CHL1, NRCAM, and neurofascin70. The conserved FIGQY motif located in the C-terminal cytosolic part of L1CAM family members is essential for binding to ankyrin-G71. Another type of cell adhesion molecule, E-cadherin, requires both ankyrin-G and β-2-spectrin for its cellular localization in early embryos and binds to ankyrin-G. Ankyrin-G interacts with the cytoplasmic tail (amino acids 738–764) of E-cadherin and provides a direct connection between E-cadherin and the spectrin/actin skeleton72. N-cadherin also expresses this highly conserved motif, and ankyrin-G binds N-cadherin and E-cadherin equally. Moreover, ankyrin-G is localized at neuromuscular junctions and costameres within skeletal muscle and binds β-dystroglycan (β-DG) and dystrophin73. Ankyrin-G binds β-DG as well as dystrophin and is required to restrict β-DG and dystrophin to costameres74. Ankyrin-G is also required for sarcolemmal integrity to organize β-DG and sarcolemmal dystrophin. The adhesion molecule CD44 interacts with 7–12 ANKRD (repeats 7–12) and binds to hyaluronic acid at its extracellular domain. This interaction can regulate prostate tumor cell transformation75 or ovarian tumor cell migration76.
Scaffold proteins/motor proteins/enzymes
The IQ motif containing J-Schwannomin-Interacting Protein 1 (IQCJ-SCHIP-1), an isoform of SCHIP-1, is highly enriched at the AIS, and its accumulation is dependent on the ANKRD of ankyrin-G77. IQCJ-SCHIP1 is associated with molecular complexes consisting of voltage-gated Nav, Kv7 channels, and cell adhesion molecules, including βIV-spectrin. IQCJ-SCHIP-1 self-associates through its C-terminus, and this association orchestrates multimolecular complexes composed of Nav/Kv7 channels, cell adhesion molecules, βIV-spectrin, and ankyrin-G in the AIS78.
Ankyrin-G associates with the anterograde motor KIF5 and Nav1.2 channel together. KIF5 binds to ankyrin-G by interacting with ANKRD 1–6 and amino acids 865–934 in the C-terminus of KIF579. Nav1.2 in the AIS was markedly reduced by the knockdown or knockout of ankyrin-G as well as the disruption of KIF5 binding to ankyrin-G through the overexpression of dominant-negative KIF5 constructs. The ankyrin-G-KIF5 complex is essential for the anterograde transport of Nav1.2 in the AIS and modulates action potential firing.
T-lymphoma invasion and metastasis 1 (Tiam1) is a guanine nucleotide exchange factor for Rho GTPases and interacts with ANKRD through amino acids 717–727 of Tiam1 (GEGTDAVKRS). Ankyrin-G binding to Tiam1 regulates the membrane localization of Tiam1 and activates a GDP/GTP exchange on Rho GTPases to regulate cytoskeletal function during cell migration80.
Recently, we found that Ubiquitin-Specific Peptidase 9 X-Linked (USP9X) modulates spine morphology through ankyrin-G stabilization2. Pathogenic USP9X variants were discovered in patients diagnosed with autistic and obsessive behaviors, delayed speech, developmental delay (DD), brain malformations, and growth retardation81,82,83. In particular, human G1890E mutation from 2 patients with ID, DD, speech delay, and ASD were found to have normal catalytic activity but impaired interaction with ankyrin-G2. The deubiquitination of proteins containing ANKRD is important for the accurate development of synapses, as a lack of USP9X results in the malfunction of synaptic structural plasticity as well as behavioral and clinical abnormalities. Notably, ankyrin-B, Shank3, and TNKS2 levels also significantly decreased in USP9X–/Y mice; male offspring that inherited the Emx1-Cre allele lacked USP9X in the telencephalon and derived cortex and hippocampal structures, indicating that USP9X stabilizes multiple ANKRD proteins in vivo.
We also reported that diacylglycerol lipase α (DAGLα) (encoded by DAGLA), which synthesizes the endocannabinoid 2-arachidonoyl-glycerol (2-AG), interacts with the ANKRD of ankyrin-G through C-terminal amino acids 709–847 (detected in a yeast-2-hybrid [Y2H] experiment). The DAGLα-ankyrin-G interaction regulates spine morphology and the lateral diffusion of DAGLα in neuronal dendrites59. Remarkably, 3 rare heterozygous variants in human DAGLA (His810Gln, Arg815His, and Ala858Val) have been identified in patients presenting with seizures and neurodevelopmental disorders, including ASD, as well as abnormalities in brain morphology84. 2-AG is a mediator of retrograde signaling to presynaptic CB1 receptors to regulate the release of neurotransmitters. However, we found that the C-terminal region of DAGLα mediated a putative nonretrograde pathway that regulates dendritic spine development via interaction with ankyrin-G.
Spectrin-binding domain
The 2 ZU5 domains and a UPA domain (ZU5N-ZU5C-UPA), followed by ANKRD, interact with spectrin/actin85. This motif has been highly conserved in ankyrin-G and across species. The DAR999AAA or Ser2417Ala mutations in ankyrin-G 480 abolished ankyrin-G binding to βII-spectrin86,87. βIV-spectrin is also known to be a component of dendritic spines. Interestingly, the DAR999AAA mutation in ankyrin-G 190 prevented its localization to the spine head, which reduced spine development and maintenance55.
Giant exon inserted domain
The neuron-specific exon 37 of ankyrin-G is inserted after the spectrin-binding domain. The resulting giant ankyrin-G transcripts are specifically expressed in the brain, and they encode ankyrin-G isoforms that migrate at 270 and 480 kDa by SDS–PAGE (ankyrin-G 270/480). The ankyrin-G 270/480 isoforms are highly enriched in AIS and are critical for organizing multiple AIS-associated proteins, including cell adhesion molecules, β-spectrins, and ion channels88. Ankyrin-G 480 interacts with GABARAP or GABARAPL1 through the Atg8 interaction motif. It regulates the stabilization of gamma-aminobutyric acid type A (GABAA) receptors at somatodendritic sites and the formation of GABAergic inhibitory synapses87,89,90. Furthermore, ankyrin-G 480 directly associates with end-binding proteins via its specific tail domain (exon 37); this interaction is crucial for AIS formation and neuronal polarity91,92. Moreover, the dynein regulator NDEL1 controls somatodendritic cargo transport at the AIS via an interaction with the specific tail domain of ankyrin-G93.
Regulatory domain
The C-terminal regulatory domain is composed of a highly conserved death domain (amino acids 1478–1562) and an unstructured stretch of 400 amino acids. Using a Y2H screen as a bait for the death domain of ankyrin-G 190, the proapoptotic molecules Fas and FADD were identified in kidney epithelial cells and confirmed by immunoprecipitation, colocalization, and pull-down assays94. This suggests that ankyrin-G may contribute to apoptotic cell death.
The EVH1 domain of Homer1b/c recognizes the PPXXF motif of ankyrin-G 190, a protein that is highly enriched in the PSD and is responsible for regulating spine morphology and maintaining a stable ankyrin-G pool in the spine head57. The ankyrin-G interaction partner DAGLα is also known to interact with Homer1 through the PPXXF motif in its C-terminal region95. These data suggest that Homer1 may regulate synaptic localization and function in complex with its binding partners within spine heads. A smaller isoform (110 kDa) of ankyrin-G lacks the membrane domain and binds directly to the p85 subunit of PI3K through both the spectrin domain and the regulatory domain of ankyrin-G. This isoform is localized to late endosomes and lysosomes and regulates signaling pathways downstream from platelet-derived growth factor receptor (PDGFR) by lysosomal-mediated degradation of tyrosine-phosphorylated PDGFR through an interaction with the p85 subunit of PI3K56.
Interactors with an unknown binding domain
Various additional interacting partners, such as GluA1, dystrophin, Hook1, and Plakophilin-2, have been found by immunoprecipitation experiments55,74,96,97, although their exact binding sites within ankyrin-G have not yet been identified.
Posttranslational modifications in ANKRD and its functional effects on ankyrin-G and binding partners
Palmitoylation
Palmitoylation is a reversible posttranscriptional modification consisting of a saturated fatty acid chain on cysteine residues through thioester bonds. This modification impacts protein hydrophobicity and allows the target protein to interact with the lipid bilayer. Genes within the zDHHC family, which includes 23 genes in humans, encode protein S-acyltransferases (PATs) that have specific interactions with their substrates and different distributions in cell compartment membranes98,99. In the brain, zDHHC proteins play various roles, including in neurite outgrowth and in synaptic plasticity, by targeting the membrane and conferring different subcellular localizations, such as to dendritic spines. Importantly, zDHHC proteins are associated with various brain disorders100,101.
To accomplish their roles as scaffold proteins, the different ankyrin-G isoforms rely on their localization in membrane microdomains102. Although several cysteines in ankyrin-G have been predicted as potential palmitoylation sites (SwissPalm), only cysteine 70 has been functionally validated103 (Fig. 3). X-ray crystallography and molecular dynamics simulations show that ankyrin-G has a preferential localization close to the membrane and that its palmitoylation stabilizes its localization to the membrane104. In epithelial cells, the absence of ankyrin-G palmitoylation prevents membrane localization under low-calcium conditions and abolishes ankyrin-G function in lateral membrane biosynthesis. In neurons, cys70 palmitoylation is important for the proper localization of ankyrin-G 270 in the AIS and the recruitment of AIS components103. This also stabilizes ankyrin-G 190 in the microdomains of dendritic spines and along the dendritic membrane105.
Ankyrin-G forms four distinct domains: an ankyrin-repeat domain (ANKRD), a spectrin-binding domain, a regulatory domain that includes a death domain (DD), and a C-terminal domain (CTD). Each domain interacts with distinct ion channels, transporters, cell adhesion proteins and signaling molecules, and cytoskeletal elements. *: PSD proteins.
A screen performed in HEK293 cells with the 23 PATs showed that zDHHC5 and zDHHC8 could palmitoylate ankyrin-G and were essential for its localization at the membrane of Madin-Darby canine kidney cells106. In neurons, only zDHHC8 was involved in ankyrin-G 190 localization in dendritic spines105, and further studies are needed to precisely define the effects of those PATs on ankyrin-G isoforms and functions. Although zDHHC5 plays an essential role in excitatory synapses107, notably through GRIP1108, few studies have linked it to SZ through GWAS109 or de novo variants3. In contrast, zDHHC8 has been identified as an SZ susceptibility gene, especially in 22q11.2 deletion syndrome110. Further studies are necessary to comprehensively understand the link between PATs and the ankyrin-G pathway in disease.
Ubiquitination
Twenty of the ANKRD proteins, which are also neuropsychiatric factors, have ubiquitinated lysine in ANKRD2. These proteins have highly conserved D boxes, which are the recognition motifs of E3 ligases. Deubiquitylating enzymes (DUBs) play key roles in stabilizing substrates by removing mono- or polyubiquitination. A recent report showed that USP9X regulates the homeostasis of ankyrin-G in early cortical neuron development2 and that this lack of USP9X causes abnormal brain morphology111. In fact, in 44 USP9X patients, 12 missense variants on the X-chromosome were identified, and these patients were reported to have speech delays and various behavioral problems83. These results indicate that the ubiquitylation signaling pathway (USP) and DUB systems coordinate various neurodevelopmental events by regulating the homeostasis of ANKRD proteins in the brain and that genetic variants can cause neurodevelopmental disorders along with various neuropsychiatric diseases.
Phosphorylation
Phosphorylation is well known to play a critical role in PPI, including in ANKRD proteins; in particular, specific phosphorylation motifs have been reported in proteins interacting with ankyrin-G (Table 1 and Fig. 4).
The ANKRD of ankyrin-G binds to cell adhesion family members (NRCAM, CHL1, Neurofascin, L1CAM) through the conserved amino acid sequence FIGQY in the cytosolic domain. The phosphorylation of the tyrosine in the FIGQY of cell adhesion proteins prevents its interaction with ankyrin-G71,112,113. EphrinB1 phosphorylates the tyrosine of cell adhesion family proteins at FIGQY via EphB and Src kinase activity, and the Src kinase inhibitor PP2 decreases the phosphorylation of L1CAM114. The tyrosine phosphorylation induced by ephrinB1 reduces the interaction with ankyrin-G, enhancing axonal growth. In contrast, the dephosphorylation of FIGQY through tyrosine phosphatases promotes ankyrin-G interactions and stabilizes synaptic terminals through a spectrin/actin cytoskeleton. Neurofascin is also known to regulate phosphorylation by activating tyrosine kinase through fibroblast growth factor-β (FGF2) or nerve growth factor or upon the enhancement of tyrosine phosphorylation through protein tyrosine phosphatase inhibitors such as dephostatin or vanadate112.
The C-terminus of RhBG is essential for its localization to the basolateral membrane of kidney epithelial cells by tyrosine phosphorylation, and dephosphorylation allows its interaction with ankyrin-G to connect to the spectrin/actin cytoskeleton65. Tyrosine 429 is phosphorylated by Src and Syk kinases. Phosphorylation is enhanced by treatment with pervanadate, a protein tyrosine phosphatase inhibitor. The phosphorylation of RhBG Tyr429 abolishes the interaction with ankyrin-G and decreases its plasma membrane stability.
The phosphorylation of voltage-gated sodium channels by the protein kinase CK2 regulates their interaction with ankyrin-G, resulting in their accumulation at the AIS. Serine sites within the conserved motif (TVTVPIAVGESDFENLNTEDFSSESDL) in the second loop of sodium channels (SCN1A, SCN2A, SCN3A, and SCN8A) are phosphorylated by CK2A, and this phosphorylation enhances their interactions with ANKRD68,115. Another auxiliary subunit of the sodium channel SCN1B undergoes tyrosine phosphorylation by FGF2 or FYN kinase in the cytosolic C-terminal region, and this phosphorylation ameliorates the interaction with ankyrin-G116. The association of ankyrin-G with Nav1 and KCNQ2/3 is altered by protein kinase CK2-mediated phosphorylation.
CK2-mediated phosphorylation of IQCJ-SCHIP-1 enhances ankyrin-G and IQCJ-SCHIP-1 interactions. Several phosphorylation sites in IQCJ-SCHIP-1 or SCHIP-1a were observed by a C-terminal tail truncation mutant of SCHIP-1, and the phosphorylation of these sites directly bound to ankyrin-G. The CK2 inhibitor 4,5,6,7-tetrabromobenzotriazole reduced IQCJ-SCHIP-1 and ankyrin-G accumulation in AIS117. The CK2-mediated IQJC-SCHIP-1 association with ankyrin-G contributed to AIS maintenance.
Forskolin treatment increases intracellular cyclic adenosine monophosphate (cAMP) levels, activating protein kinase A (PKA) signaling and initiating a signaling cascade leading to DAGLα serine 738 phosphorylation. In this signaling mechanism, the phosphorylation of serine 738 is inhibited by treatment with RP-8-Br-cAMPS, a PKA inhibitor. Forskolin treatment-induced serine 738 phosphorylation of DAGLα enhances its interaction with ankyrin-G, increasing spine size and decreasing DAGLα surface diffusion59. These results suggest that cAMP-mediated acute spine enlargement is regulated through an independent retrograde endogenous cannabinoid system59.
USP9X is a key regulator of the transforming growth factor-β (TGFβ) signaling pathway118, regulating dendritic development and neuronal axonal growth in response to TGFβ stimulation119. TGFβ receptors, or serine/threonine kinase receptors, activate downstream signaling cascades such as the extracellular signal-regulated kinase, phosphoinositide 3-kinase, mitogen-activated protein kinase, Smad family, AKT, nuclear factor-kB, and JUN N-terminal kinase cascades120. TGFβ treatment phosphorylates serine 1593 and 1609 in USP9X, and these modifications enhance the interaction with ankyrin-G to modulate spine morphology2,121. TGFβ treatment also phosphorylates serine 1600 and regulates USP9X activity rather than binding to ankyrin-G122. In addition, insulin is known to phosphorylate serine 1593 and is inhibited by MK-2206 (Akt1/2/3 inhibitor)123. This suggests that DUBs may be an ideal drug target for the enhancement or inhibition of substrate binding124. Recently, an optogenetic system (optoTGFBRs) was engineered to precisely control TGF-β signaling temporally and spatially125. Using this system, it is possible to selectively control signal activity through light stimulation of specific cells or tissues at the desired time. In the future, these techniques may be useful in developing therapeutic approaches to treat human psychiatric disorders.
Conclusion
ANKRD proteins are structurally capable of performing various intracellular physiological functions by binding to partner proteins. In particular, several isoforms of ankyrin-G in the brain are formed through alternative splicing and play significant roles in different intracellular locations. In addition, these isoforms are expressed quantitatively at different developmental periods to regulate neuronal development. The ankyrin-G-related PPI network is significantly involved in neuropsychiatric diseases such as ASD, BD, and SZ. These results suggest that the modification of ankyrin-G and related proteins is a potential therapeutic approach for neuropsychiatric disorders. In the future, further studies should be performed to increase our understanding of these detailed mechanisms, and they will be helpful in the development of personalized treatments for individual patients with fewer side effects.
References
Mosavi, L. K., Cammett, T. J., Desrosiers, D. C. & Peng, Z. Y. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 13, 1435–1448 (2004).
Yoon, S. et al. Usp9X controls ankyrin-repeat domain protein homeostasis during dendritic spine development. Neuron 105, 506–521 e507 (2020).
Fromer, M. et al. De novo mutations in schizophrenia implicate synaptic networks. Nature 506, 179–184 (2014).
Iossifov, I. et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221 (2014).
Purcell, S. M. et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature 506, 185–190 (2014).
Hou, L. et al. Genome-wide association study of 40,000 individuals identifies two novel loci associated with bipolar disorder. Hum. Mol. Genet. 25, 3383–3394 (2016).
Mullins, N. et al. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat. Genet. 53, 817–829 (2021).
Ferreira, M. A. et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat. Genet. 40, 1056–1058 (2008).
Schulze, T. G. et al. Two variants in Ankyrin 3 (ANK3) are independent genetic risk factors for bipolar disorder. Mol. Psychiatry 14, 487–491 (2009).
Sanders, S. J. et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 485, 237–241 (2012).
Kloth, K. et al. First de novo ANK3 nonsense mutation in a boy with intellectual disability, speech impairment and autistic features. Eur. J. Med. Genet. 60, 494–498 (2017).
Baum, A. E. et al. Meta-analysis of two genome-wide association studies of bipolar disorder reveals important points of agreement. Mol. Psychiatry 13, 466–467 (2008).
Ruberto, G. et al. The cognitive impact of the ANK3 risk variant for bipolar disorder: initial evidence of selectivity to signal detection during sustained attention. PLoS ONE 6, e16671 (2011).
Hatzimanolis, A. et al. Bipolar disorder ANK3 risk variant effect on sustained attention is replicated in a large healthy population. Psychiatr. Genet. 22, 210–213 (2012).
Linke, J. et al. Genome-wide supported risk variant for bipolar disorder alters anatomical connectivity in the human brain. Neuroimage 59, 3288–3296 (2012).
Delvecchio, G., Dima, D. & Frangou, S. The effect of ANK3 bipolar-risk polymorphisms on the working memory circuitry differs between loci and according to risk-status for bipolar disorder. Am. J. Med. Genet. B Neuropsychiatr. Genet. 168B, 188–196 (2015).
Hori, H. et al. Cognitive effects of the ANK3 risk variants in patients with bipolar disorder and healthy individuals. J. Affect. Disord. 158, 90–96 (2014).
Dima, D. et al. Independent modulation of engagement and connectivity of the facial network during affect processing by CACNA1C and ANK3 risk genes for bipolar disorder. JAMA Psychiatry 70, 1303–1311 (2013).
Ota, M. et al. Effects of ankyrin 3 gene risk variants on brain structures in patients with bipolar disorder and healthy subjects. Psychiat Clin. Neurosci. 70, 498–506 (2016).
Lippard, E. T. C. et al. Effects of ANK3 variation on gray and white matter in bipolar disorder. Mol. Psychiatry 22, 1345–1351 (2017).
Wirgenes, K. V. et al. ANK3 gene expression in bipolar disorder and schizophrenia. Br. J. Psychiatry 205, 244–245 (2014).
Rueckert, E. H. et al. Cis-acting regulation of brain-specific ANK3 gene expression by a genetic variant associated with bipolar disorder. Mol. Psychiatry 18, 922–929 (2013).
Tang, L. L. et al. ANK3 gene polymorphism Rs10994336 influences executive functions by modulating methylation in patients with bipolar disorder. Front. Neurosci. 15, 682873 (2021).
Hughes, T. et al. Elevated expression of a minor isoform of ANK3 is a risk factor for bipolar disorder. Transl. Psychiatry 8, 210 (2018).
Hughes, T. et al. A loss-of-function variant in a minor isoform of ANK3 protects against bipolar disorder and schizophrenia. Biol. Psychiatry 80, 323–330 (2016).
Bavamian, S. et al. Dysregulation of miR-34a links neuronal development to genetic risk factors for bipolar disorder. Mol. Psychiatry 20, 573–584 (2015).
Bame, M., McInnis, M. G. & O’Shea, K. S. MicroRNA alterations in induced pluripotent stem cell-derived neurons from bipolar disorder patients: pathways involved in neuronal differentiation, axon guidance, and plasticity. Stem Cells Dev. 29, 1145–1159 (2020).
Fiorentino, A. et al. Analysis of ANK3 and CACNA1C variants identified in bipolar disorder whole genome sequence data. Bipolar Disord. 16, 583–591 (2014).
Forstner, A. J. et al. Whole-exome sequencing of 81 individuals from 27 multiply affected bipolar disorder families. Transl. Psychiatry 10, 57 (2020).
Nelson, A. D. et al. Ankyrin-G regulates forebrain connectivity and network synchronization via interaction with GABARAP. Mol. Psychiatry 25, 2800–2817 (2020).
Gella, A. et al. Is Ankyrin a genetic risk factor for psychiatric phenotypes? BMC Psychiatry 11, 103 (2011).
Athanasiu, L. et al. Gene variants associated with schizophrenia in a Norwegian genome-wide study are replicated in a large European cohort. J. Psychiatr. Res. 44, 748–753 (2010).
Moskvina, V. et al. Gene-wide analyses of genome-wide association data sets: evidence for multiple common risk alleles for schizophrenia and bipolar disorder and for overlap in genetic risk. Mol. Psychiatry 14, 252–260 (2009).
Yuan, A. H. et al. ANK3 as a risk gene for schizophrenia: new data in Han Chinese and meta analysis. Am. J. Med. Genet. B 159b, 997–1005 (2012).
Lim, C. H. et al. Genetic association of LMAN2L gene in schizophrenia and bipolar disorder and its interaction with ANK3 gene polymorphism. Prog. Neuropsychopharmacol. Biol. Psychiatry 54, 157–162 (2014).
Nie, F. et al. Genetic analysis of SNPs in CACNA1C and ANK3 gene with schizophrenia: a comprehensive meta-analysis. Am. J. Med. Genet. B Neuropsychiatr. Genet. 168, 637–648 (2015).
Guo, X. J. et al. Association analysis of ANK3 gene variants with schizophrenia in a northern Chinese Han population. Oncotarget 7, 85888–85894 (2016).
Roussos, P. et al. Molecular and genetic evidence for abnormalities in the nodes of Ranvier in schizophrenia. Arch. Gen. Psychiat. 69, 7–15 (2012).
Zhong, X., Zhang, L., Han, S., An, Z. & Yi, Q. Case control study of association between the ANK3 rs10761482 polymorphism and schizophrenia in persons of Uyghur nationality living in Xinjiang China. Shanghai Arch. Psychiatry 26, 288–293 (2014).
Roussos, P., Giakoumaki, S. G., Georgakopoulos, A., Robakis, N. K. & Bitsios, P. The CACNA1C and ANK3 risk alleles impact on affective personality traits and startle reactivity but not on cognition or gating in healthy males. Bipolar Disord. 13, 250–259 (2011).
Cassidy, C. et al. Association of a risk allele of ANK3 with cognitive performance and cortical thickness in patients with first-episode psychosis. J. Psychiatr. Neurosci. 39, 31–39 (2014).
Lam, M. et al. Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat. Genet. 51, 1670–1678 (2019).
Legge, S. E. et al. Association of genetic liability to psychotic experiences with neuropsychotic disorders and traits. JAMA Psychiatry 76, 1256–1265 (2019).
Bi, C. et al. Mutations of ANK3 identified by exome sequencing are associated with autism susceptibility. Hum. Mutat. 33, 1635–1638 (2012).
Codina-Sola, M. et al. Integrated analysis of whole-exome sequencing and transcriptome profiling in males with autism spectrum disorders. Mol. Autism 6, 21 (2015).
Bonnet-Brilhault, F. et al. GABA/Glutamate synaptic pathways targeted by integrative genomic and electrophysiological explorations distinguish autism from intellectual disability. Mol. Psychiatry 21, 411–418 (2016).
Chen, S., Xing, Y. & Kang, J. Latent and abnormal functional connectivity circuits in autism spectrum disorder. Front. Neurosci. 11, 125 (2017).
Lim, E. T. et al. Rates, distribution and implications of postzygotic mosaic mutations in autism spectrum disorder. Nat. Neurosci. 20, 1217–1224 (2017).
McKenna, B., Koomar, T., Vervier, K., Kremsreiter, J. & Michaelson, J. J. Whole-genome sequencing in a family with twin boys with autism and intellectual disability suggests multimodal polygenic risk. Cold Spring Harb. Mol. Case Stud. 4, a003285 (2018).
Feliciano, P. et al. Exome sequencing of 457 autism families recruited online provides evidence for autism risk genes. NPJ Genom. Med. 4, 19 (2019).
Kloth, K. et al. ANK3 related neurodevelopmental disorders: expanding the spectrum of heterozygous loss-of-function variants. Neurogenetics 22, 263–269 (2021).
Zhou, D. X. et al. Ankyrin(G) is required for clustering of voltage-gated Na channels at axon initial segments and for normal action potential firing. J. Cell Biol. 143, 1295–1304 (1998).
Leussis, M. P. et al. The ANK3 bipolar disorder gene regulates psychiatric-related behaviors that are modulated by lithium and stress. Biol. Psychiatry 73, 683–690 (2013).
Chang, K. J. et al. Glial ankyrins facilitate paranodal axoglial junction assembly. Nat. Neurosci. 17, 1673–1681 (2014).
Smith, K. R. et al. Psychiatric risk factor ANK3/ankyrin-G nanodomains regulate the structure and function of glutamatergic synapses. Neuron 84, 399–415 (2014).
Ignatiuk, A., Quickfall, J. P., Hawrysh, A. D., Chamberlain, M. D. & Anderson, D. H. The smaller isoforms of ankyrin 3 bind to the p85 subunit of phosphatidylinositol 3’-kinase and enhance platelet-derived growth factor receptor down-regulation. J. Biol. Chem. 281, 5956–5964 (2006).
Yoon, S. et al. Homer1 promotes dendritic spine growth through ankyrin-G and its loss reshapes the synaptic proteome. Mol. Psychiatry 26, 1775–1789 (2021).
Zhu, S. et al. Genetic disruption of ankyrin-G in adult mouse forebrain causes cortical synapse alteration and behavior reminiscent of bipolar disorder. Proc. Natl Acad. Sci. USA 114, 10479–10484 (2017).
Yoon, S., Myczek, K. & Penzes, P. cAMP signaling-mediated phosphorylation of diacylglycerol lipase alpha regulates interaction with ankyrin-G and dendritic spine morphology. Biol. Psychiatry 90, 263–274 (2021).
Cruz, D. A., Lovallo, E. M., Stockton, S., Rasband, M. & Lewis, D. A. Postnatal development of synaptic structure proteins in pyramidal neuron axon initial segments in monkey prefrontal cortex. J. Comp. Neurol. 514, 353–367 (2009).
Yoshimura, T., Stevens, S. R., Leterrier, C., Stankewich, M. C. & Rasband, M. N. Developmental changes in expression of betaIV spectrin splice variants at axon initial segments and nodes of Ranvier. Front. Cell. Neurosci. 10, 304 (2016).
Kizhatil, K., Baker, S. A., Arshavsky, V. Y. & Bennett, V. Ankyrin-G promotes cyclic nucleotide-gated channel transport to rod photoreceptor sensory cilia. Science 323, 1614–1617 (2009).
Zhang, X., Davis, J. Q., Carpenter, S. & Bennett, V. Structural requirements for association of neurofascin with ankyrin. J. Biol. Chem. 273, 30785–30794 (1998).
Lopez, C. et al. The ammonium transporter RhBG: requirement of a tyrosine-based signal and ankyrin-G for basolateral targeting and membrane anchorage in polarized kidney epithelial cells. J. Biol. Chem. 280, 8221–8228 (2005).
Sohet, F. et al. Phosphorylation and ankyrin-G binding of the C-terminal domain regulate targeting and function of the ammonium transporter RhBG. J. Biol. Chem. 283, 26557–26567 (2008).
Genetet, S., Ripoche, P., Le Van Kim, C., Colin, Y. & Lopez, C. Evidence of a structural and functional ammonium transporter RhBG.anion exchanger 1.ankyrin-G complex in kidney epithelial cells. J. Biol. Chem. 290, 6925–6936 (2015).
Garrido, J. J. et al. A targeting motif involved in sodium channel clustering at the axonal initial segment. Science 300, 2091–2094 (2003).
Lemaillet, G., Walker, B. & Lambert, S. Identification of a conserved ankyrin-binding motif in the family of sodium channel alpha subunits. J. Biol. Chem. 278, 27333–27339 (2003).
Pan, Z. et al. A common ankyrin-G-based mechanism retains KCNQ and NaV channels at electrically active domains of the axon. J. Neurosci. 26, 2599–2613 (2006).
Bennett, V. & Lorenzo, D. N. An adaptable spectrin/ankyrin-based mechanism for long-range organization of plasma membranes in vertebrate tissues. Curr. Top. Membr. 77, 143–184 (2016).
Jenkins, S. M. et al. FIGQY phosphorylation defines discrete populations of L1 cell adhesion molecules at sites of cell-cell contact and in migrating neurons. J. Cell Sci. 114, 3823–3835 (2001).
Kizhatil, K. et al. Ankyrin-G is a molecular partner of E-cadherin in epithelial cells and early embryos. J. Biol. Chem. 282, 26552–26561 (2007).
Kordeli, E., Ludosky, M. A., Deprette, C., Frappier, T. & Cartaud, J. Ankyrin(G) is associated with the postsynaptic membrane and the sarcoplasmic reticulum in the skeletal muscle fiber. J. Cell Sci. 111, 2197–2207 (1998).
Ayalon, G., Davis, J. Q., Scotland, P. B. & Bennett, V. An ankyrin-based mechanism for functional organization of dystrophin and dystroglycan. Cell 135, 1189–1200 (2008).
Zhu, D. & Bourguignon, L. Y. W. The ankyrin-binding domain of CD44s is involved in regulating hyaluronic acid-mediated functions and prostate tumor cell transformation. Cell Motil. Cytoskeleton 39, 209–222 (1998).
Zhu, D. & Bourguignon, L. Y. W. Interaction between CD44 and the repeat domain of ankyrin promotes hyaluronic acid-mediated ovarian tumor cell migration. J. Cell. Physiol. 183, 182–195 (2000).
Martin, P. M. et al. Schwannomin-interacting protein-1 isoform IQCJ-SCHIP-1 is a late component of nodes of Ranvier and axon initial segments. J. Neurosci. 28, 6111–6117 (2008).
Martin, P. M. et al. Schwannomin-interacting protein 1 isoform IQCJ-SCHIP1 is a multipartner ankyrin- and spectrin-binding protein involved in the organization of nodes of Ranvier. J. Biol. Chem. 292, 2441–2456 (2017).
Barry, J. et al. Ankyrin-G directly binds to kinesin-1 to transport voltage-gated Na+ channels into axons. Dev. Cell 28, 117–131 (2014).
Bourguignon, L. Y., Zhu, H., Shao, L. & Chen, Y. W. Ankyrin-Tiam1 interaction promotes Rac1 signaling and metastatic breast tumor cell invasion and migration. J. Cell Biol. 150, 177–191 (2000).
Homan, C. C. et al. Mutations in USP9X are associated with X-linked intellectual disability and disrupt neuronal cell migration and growth. Am. J. Hum. Genet. 94, 470–478 (2014).
Reijnders, M. R. et al. De novo loss-of-function mutations in USP9X cause a female-specific recognizable syndrome with developmental delay and congenital malformations. Am. J. Hum. Genet. 98, 373–381 (2016).
Johnson, B. V. et al. Partial loss of USP9X function leads to a male neurodevelopmental and behavioral disorder converging on transforming growth factor beta signaling. Biol. Psychiatry 87, 100–112 (2020).
Smith, D. R., Stanley, C. M., Foss, T., Boles, R. G. & McKernan, K. Rare genetic variants in the endocannabinoid system genes CNR1 and DAGLA are associated with neurological phenotypes in humans. PLoS ONE 12, e0187926 (2017).
Wang, C., Yu, C., Ye, F., Wei, Z. & Zhang, M. Structure of the ZU5-ZU5-UPA-DD tandem of ankyrin-B reveals interaction surfaces necessary for ankyrin function. Proc. Natl Acad. Sci. USA 109, 4822–4827 (2012).
Kizhatil, K. et al. Ankyrin-G and beta2-spectrin collaborate in biogenesis of lateral membrane of human bronchial epithelial cells. J. Biol. Chem. 282, 2029–2037 (2007).
Jenkins, P. M. et al. Giant ankyrin-G: a critical innovation in vertebrate evolution of fast and integrated neuronal signaling. Proc. Natl Acad. Sci. USA 112, 957–964 (2015).
Rasband, M. N. The axon initial segment and the maintenance of neuronal polarity. Nat. Rev. Neurosci. 11, 552–562 (2010).
Tseng, W. C., Jenkins, P. M., Tanaka, M., Mooney, R. & Bennett, V. Giant ankyrin-G stabilizes somatodendritic GABAergic synapses through opposing endocytosis of GABAA receptors. Proc. Natl Acad. Sci. USA 112, 1214–1219 (2015).
Li, J. et al. Potent and specific Atg8-targeting autophagy inhibitory peptides from giant ankyrins. Nat. Chem. Biol. 14, 778–787 (2018).
Leterrier, C. et al. End-binding proteins EB3 and EB1 link microtubules to ankyrin G in the axon initial segment. Proc. Natl Acad. Sci. USA 108, 8826–8831 (2011).
Freal, A. et al. Cooperative interactions between 480 kDa ankyrin-G and EB proteins assemble the axon initial segment. J. Neurosci. 36, 4421–4433 (2016).
Kuijpers, M. et al. Dynein regulator NDEL1 controls polarized cargo transport at the axon initial segment. Neuron 89, 461–471 (2016).
Del Rio, M. et al. The death domain of kidney ankyrin interacts with Fas and promotes Fas-mediated cell death in renal epithelia. J. Am. Soc. Nephrol. 15, 41–51 (2004).
Jung, K. M. et al. A key role for diacylglycerol lipase-alpha in metabotropic glutamate receptor-dependent endocannabinoid mobilization. Mol. Pharmacol. 72, 612–621 (2007).
Weimer, J. M., Chattopadhyay, S., Custer, A. W. & Pearce, D. A. Elevation of Hook1 in a disease model of Batten disease does not affect a novel interaction between Ankyrin G and Hook1. Biochem. Biophys. Res. Commun. 330, 1176–1181 (2005).
Sato, P. Y. et al. Interactions between ankyrin-G, Plakophilin-2, and Connexin43 at the cardiac intercalated disc. Circ. Res. 109, 193–201 (2011).
Fukata, M., Fukata, Y., Adesnik, H., Nicoll, R. A. & Bredt, D. S. Identification of PSD-95 palmitoylating enzymes. Neuron 44, 987–996 (2004).
Lemonidis, K. et al. The zDHHC family of S-acyltransferases. Biochem. Soc. Trans. 43, 217–221 (2015).
Globa, A. K. & Bamji, S. X. Protein palmitoylation in the development and plasticity of neuronal connections. Curr. Opin. Neurobiol. 45, 210–220 (2017).
Sanders, S. S. et al. Curation of the mammalian palmitoylome indicates a pivotal role for palmitoylation in diseases and disorders of the nervous system and cancers. PLoS Comput. Biol. 11, e1004405 (2015).
Smith, K. R. & Penzes, P. Ankyrins: roles in synaptic biology and pathology. Mol. Cell. Neurosci. 91, 131–139 (2018).
He, M., Jenkins, P. & Bennett, V. Cysteine 70 of ankyrin-G is S-palmitoylated and is required for function of ankyrin-G in membrane domain assembly. J. Biol. Chem. 287, 43995–44005 (2012).
Fujiwara, Y. et al. Structural basis for the membrane association of ankyrinG via palmitoylation. Sci. Rep. 6, 23981 (2016).
Piguel, N. H. et al. The 190 kDa Ankyrin-G isoform is required for the dendritic stability of neurons and its palmitoylation is altered by lithium. bioRxiv https://doi.org/10.1101/620708 (2019).
He, M., Abdi, K. M. & Bennett, V. Ankyrin-G palmitoylation and betaII-spectrin binding to phosphoinositide lipids drive lateral membrane assembly. J. Cell Biol. 206, 273–288 (2014).
Shimell, J. J. et al. Regulation of hippocampal excitatory synapses by the Zdhhc5 palmitoyl acyltransferase. J. Cell Sci. 134, 254276 (2021).
Thomas, G. M., Hayashi, T., Chiu, S. L., Chen, C. M. & Huganir, R. L. Palmitoylation by DHHC5/8 targets GRIP1 to dendritic endosomes to regulate AMPA-R trafficking. Neuron 73, 482–496 (2012).
Zhao, Y. et al. Integrating genome-wide association study and expression quantitative trait locus study identifies multiple genes and gene sets associated with schizophrenia. Prog. Neuropsychopharmacol. Biol. Psychiatry 81, 50–54 (2018).
Qin, X., Chen, J. & Zhou, T. 22q11.2 deletion syndrome and schizophrenia. Acta Biochim. Biophys. Sin. 52, 1181–1190 (2020).
Oishi, S. et al. Usp9x-deficiency disrupts the morphological development of the postnatal hippocampal dentate gyrus. Sci. Rep. 6, 25783 (2016).
Garver, T. D., Ren, Q., Tuvia, S. & Bennett, V. Tyrosine phosphorylation at a site highly conserved in the L1 family of cell adhesion molecules abolishes ankyrin binding and increases lateral mobility of neurofascin. J. Cell Biol. 137, 703–714 (1997).
Tuvia, S., Garver, T. D. & Bennett, V. The phosphorylation state of the FIGQY tyrosine of neurofascin determines ankyrin-binding activity and patterns of cell segregation. Proc. Natl Acad. Sci. USA 94, 12957–12962 (1997).
Dai, J. et al. EphB regulates L1 phosphorylation during retinocollicular mapping. Mol. Cell. Neurosci. 50, 201–210 (2012).
Brechet, A. et al. Protein kinase CK2 contributes to the organization of sodium channels in axonal membranes by regulating their interactions with ankyrin G. J. Cell Biol. 183, 1101–1114 (2008).
Lux, S. E., John, K. M. & Bennett, V. Analysis of cDNA for human erythrocyte ankyrin indicates a repeated structure with homology to tissue-differentiation and cell-cycle control proteins. Nature 344, 36–42 (1990).
Papandreou, M. J. et al. CK2-regulated schwannomin-interacting protein IQCJ-SCHIP-1 association with AnkG contributes to the maintenance of the axon initial segment. J. Neurochem. 134, 527–537 (2015).
Dupont, S. et al. FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell 136, 123–135 (2009).
Stegeman, S. et al. Loss of Usp9x disrupts cortical architecture, hippocampal development and TGFbeta-mediated axonogenesis. PLoS ONE 8, e68287 (2013).
Akhurst, R. J. & Hata, A. Targeting the TGFbeta signalling pathway in disease. Nat. Rev. Drug Discov. 11, 790–811 (2012).
Yoon, S., Parnell, E. & Penzes, P. TGF-beta-induced phosphorylation of Usp9X stabilizes Ankyrin-G and regulates dendritic spine development and maintenance. Cell Rep. 31, 107685 (2020).
Naik, E. & Dixit, V. M. Usp9X is required for lymphocyte activation and homeostasis through its control of ZAP70 ubiquitination and PKCbeta kinase activity. J. Immunol. 196, 3438–3451 (2016).
Humphrey, S. J. et al. Dynamic adipocyte phosphoproteome reveals that Akt directly regulates mTORC2. Cell Metab. 17, 1009–1020 (2013).
Kane, E. I. & Spratt, D. E. Structural insights into ankyrin repeat-containing proteins and their influence in ubiquitylation. Int. J. Mol. Sci. 22, 609 (2021).
Li, Y. et al. Spatiotemporal control of TGF-beta signaling with light. ACS Synth. Biol. 7, 443–451 (2018).
Acknowledgements
Figure 4 was created with Biorender.com (https://biorender.com/). Special thanks to Dr. Marc Forrest and Dr. Euan Parnell for proofreading.
Funding
This review was supported by NIH grants R01MH107182 and R01MH097216.
Author information
Authors and Affiliations
Contributions
S.Y. and N.P. collected information and wrote the article. P.P. revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yoon, S., Piguel, N.H. & Penzes, P. Roles and mechanisms of ankyrin-G in neuropsychiatric disorders. Exp Mol Med 54, 867–877 (2022). https://doi.org/10.1038/s12276-022-00798-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s12276-022-00798-w
This article is cited by
-
ANK3 rs10994336 and ZNF804A rs7597593 polymorphisms: genetic interaction for emotional and behavioral symptoms of alcohol withdrawal syndrome
BMC Psychiatry (2024)
-
Neurofilament accumulations in amyotrophic lateral sclerosis patients’ motor neurons impair axonal initial segment integrity
Cellular and Molecular Life Sciences (2023)