Abstract
Total knee arthroplasty (TKA) is the most common and cost-effective treatment for older adults with long-standing osteoarthritis. During TKA, muscle cells suffer from prolonged oxygen deficiency, which leads to altered cell metabolism that reduces the energy demand and maintains cell homeostasis before blood flow is restored. This study focused on the role of the lncRNA muscleblind-like 1 antisense RNA 1 (MBNL1-AS1) in protecting sevoflurane-pretreated mice against ischemia-reperfusion (I/R) injury after TKA, as well as the elucidation of the potential associated mechanism. Identification of differentially expressed lncRNAs was performed using the microarray dataset GSE21164, which was extracted from the GEO database. Target genes of the lncRNA were determined using Multi-Experiment Matrix (MEM), a dual-luciferase reporter gene assay, and KEGG enrichment analyses. The results showed that MBNL1-AS1 was overexpressed in skeletal muscle cells in mice, while KCNMA1, which was enriched in the cGMP-PKG signaling pathway, was negatively regulated by MBNL1-AS1. Furthermore, I/R mice displayed serious inflammatory reactions. Down-regulation of MBNL1-AS1 increased the expression of KCNMA1, PKGII, VASP, VEGF, Bcl-2, Cyclin D1, Cyclin D3, and Cdc 42 but decreased the expression of Bax, cleaved caspase-3, and cleaved PARP. Furthermore, upon MBNL1-AS1 upregulation, the rate of cell apoptosis increased while the rate of cell proliferation decreased. Our data suggested that down-regulated lncRNA MBNL1-AS1 might promote the proliferation and inhibit the apoptosis of skeletal muscle cells by upregulating KCNMA1 expression via activation of the cGMP-PKG signaling pathway, thus protecting sevoflurane-pretreated mice against I/R injury after TKA.
Similar content being viewed by others
Introduction
Total knee arthroplasty (TKA) is one of the most common orthopedic operations in elderly patients with end-stage osteoarthritis (OA), a degenerative disease greatly affecting the quality of life of older adults1. TKA has been widely used in China due to its excellent mid-term prognosis2. During orthopedic procedures such as TKA, tourniquets are generally applied to control blood loss and to maintain a relatively clear surgical field3. However, when tourniquets are used, skeletal muscles can suffer from ischemia, while subsequent reperfusion into ischemic muscles can cause severe complications4. Most importantly, ischemia-reperfusion (I/R) injury caused by the application of tourniquets can lead to oxidative muscle injury and muscle atrophy, which can hinder full functional recovery after TKA surgery. Nevertheless, how I/R injury affects the functions of human skeletal muscles at the cellular level remains poorly understood5. Furthermore, increasing evidence has demonstrated the beneficial role of sevoflurane (Sevo) against I/R injury6,7. In this study, we aimed to explore the molecular mechanisms underlying the protective role of Sevo against I/R injury after TKA.
Gene Expression Omnibus (GEO) is a public repository database for high-throughput gene sequencing, hybridization arrays, chips, and microarrays8. A previous study reported the identification of differentially expressed lncRNAs using the GSE21164 microarray dataset from the GEO database9. In the present study, muscleblind-like 1 antisense RNA 1 (MBNL1-AS1) was determined to be a differentially expressed lncRNA implicated in I/R injury after TKA. Based on data obtained from the Genotype-Tissue Expression (GTEx) project, a database on the variation of gene expression in human tissues10, a correlation (log2-scale) was found between MBNL1-AS1 and potassium calcium-activated channel subfamily M alpha 1 (KCNMA1). Interestingly, in the present study, MBNL1-AS1 was also identified as a putative target gene of KCNMA1 based on the Multi-Experiment Matrix (MEM; http://biit.cs.ut.ee/mem/), a web resource of gene co-expression data arranged by different species and types of microarray platforms11. Furthermore, previous Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses based on the WebGestalt database indicated that KCNMA1 participates in the cyclic guanosine monophosphate-protein kinase G (cGMP-PKG) signaling pathway, which plays critical roles in cellular functions12,13,14. Therefore, this study aimed to explore the role of MBNL1-AS1 in protecting the skeletal muscle cells of Sevo-pretreated mice against I/R injury after TKA and the role of KCNMA1 and the cGMP-PKG signaling pathway in the underlying molecular mechanism of MBNL1-AS1.
Materials and methods
Ethics statement
The experimental protocol and animal use plan in this study were approved by the Animal Ethics Committee of China-Japan Union Hospital of Jilin University.
Bioinformatics prediction
The GSE21164 microarray dataset and probe files for TKA downloaded from the GEO database (http://www.ncbi.nlm.nih.gov/geo) were analyzed using the Affymetrix Human Genome U133 Plus 2.0 Array (Affymetrix, Santa Clara, CA). Background correction and normalization of GSE21164 were performed with the Affy installation package in R software15. Identification of differentially expressed lncRNAs in GSE21164 was performed using an empirical Bayes method in conjunction with a linear model and t-test16. Putative target genes of lncRNA were determined based on MEM (http://biit.cs.ut.ee/mem/). The target genes enriched in the cGMP-PKG pathway were ascertained using the KEGG pathway database and the WebGestalt database (http://www.webgestalt.org)17.
Experimental animals and model establishment
A total of 60 healthy male Kunming mice (weight: 30 ± 2 g; age: 6–8 weeks) were obtained from the Institute of Laboratory Animal Science, Chinese Academy of Medical Sciences (Beijing, China). All mice were kept under specific temperature (18–22 °C), relative humidity (40–70%), and noise (<50 dB) conditions, with ad libitum access to water and food in an animal room with a 12-h light/dark cycle. The mice were randomly assigned to a normal group, an I/R group and a Sevo group (I/R mice treated with Sevo after TKA), with 20 mice in each group.
During model establishment, the mice were fixed on a super-clean table, anesthetized by intraperitoneal injection of pentobarbital (30 mg/kg) and disinfected. Subsequently, an incision along the midline of the knee was opened through the skin, subcutaneous tissue, and articular capsule of each mouse. The articular capsule was then opened to expose the right knee joint. Next, a 1-mm section of articular cartilage was removed from the distal femur using an electric drill. A sterile prosthesis was then inserted into the intramedullary cavity of the femur using a homemade pointed cone. The incision was then cleaned and sutured. The mice were given intramuscular injection of gentamycin (200 U) continuously for 3 days. After 2 weeks of normal feeding, I/R was performed.
Before I/R, the mice in the Sevo group received 3 doses of 2.0% Sevo (0426, Fuso Pharmaceutical Industries, Ltd., Osaka, Japan) at 15 min intervals from a Sevo vaporizer (Draeger Medical, Lubeck, Germany).
Mice in the I/R and Sevo groups were then fixed on the operating table in a supine position, anesthetized by intraperitoneal injection of pentobarbital (30 mg/kg) and disinfected. Their femoral arteries were fixed and separated. An atraumatic artery clip was used to block the artery in each mouse for 5 h, and blood flow was restored 12 h later.
Two weeks later, the mice were anesthetized by intraperitoneal injection of 2% pentobarbital sodium. Subsequently, the knee joint cartilage was removed from each mouse and washed in ice-cold saline to remove the blood. After shearing and grinding, the knee joint cartilage was centrifuged at 3000 r/min for 15 min to prepare a tissue homogenate. Next, the supernatant was collected and stored in a 1.5-mL Eppendorf tube at −20 °C. At the same time, a section of the limb skeletal muscle was collected and stored in liquid nitrogen, while the remaining limb skeletal muscle was fixed in 10% formaldehyde, dehydrated in gradient alcohol for 24 h, and embedded in paraffin.
Hematoxylin–eosin (HE) staining
Skeletal muscle tissues collected from four mice in each group were sliced into 5 μm serial sections, followed by extension at 45 °C and 1 h drying at 60 °C. After being dewaxed in xylene and rehydrated, the sections were stained with hematoxylin (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) for 2 min, washed in tap water for 10 s, incubated for 10 s in alcohol containing 1% hydrochloric acid, and washed for 1 min in distilled water. Subsequently, the sections were counterstained with eosin and washed for 10 s in distilled water, followed by dehydration in gradient alcohol. Finally, the sections were cleared in xylene and mounted in neutral gum. Histological changes in skeletal muscle tissues were observed under an optical microscope (XP-330, Shanghai Bingyu Optical Instrument Co., Ltd., Shanghai, China).
Enzyme-linked immunosorbent assay (ELISA)
The concentrations of lactic dehydrogenase (LDH) and inflammatory factors, such as tumor necrosis factor-α (TNF-α) and interleukin-1β (IL-1β), in plasma were determined using corresponding ELISA kits (LDH kit, No: 59-20044, TNF-α kit, No: 69-98069, and IL-1β kit, No: 69-98070). ELISA assays were carried out by strictly following the procedures provided by the manufacturer (Wuhan Moshake Biological Technology Co., Ltd., Wuhan, China).
Immunohistochemistry assay
Sections were dewaxed twice in xylene (10 min each time) and dehydrated in graded ethanol (100, 95, 80, and 70%, 2 min each time). Subsequently, the sections were placed on a shaker and washed twice in distilled water (5 min each time). Next, the sections were incubated in 3% H2O2 for 10 min and washed with distilled water, followed by antigen repair at high pressure for 90 s. After being cooled to room temperature, the sections were washed with phosphate buffer saline (PBS), blocked with 5% bovine serum albumin (BSA) for 30 min at 37 °C, and then incubated overnight at 4 °C with rabbit anti-mouse KCNMA1 primary antibodies (1:100, CSB-EP012077PI, Cusabio Biotech Co., Ltd., Wuhan, China). After PBS washing, the sections were then incubated with biotinylated secondary antibodies (goat anti-rabbit immunoglobulin G (IgG); 1:100, SF8-0.3, Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) at 37 °C for 30 min. Subsequently, after being fully washed, the sections were incubated with horseradish peroxidase (HRP)-conjugated working fluid, stained with 3,3-diaminobenzidine (DAB, chromogenic agent), and counterstained for 5 min with hematoxylin. During this experiment, PBS was used as the negative control. After staining, cells showing a brownish yellow membrane or cytoplasm were determined to be KCNMA1 positive. Five fields of vision were chosen randomly for each section under a high-power lens to detect the localization of positive expression in skeletal muscle cells. The proportion of positive cells was determined to be the positive expression rate. The experiment was conducted 3 times.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Skeletal muscle tissues were treated with 1 mL TRIzol® reagent (Invitrogen Inc., Carlsbad, CA, USA) and grinded in an ice bath to extract total RNA according to the manufacturer’s protocol. The purity and concentration of extracted RNA were measured by ultraviolet (UV) spectrophotometry (UV1901, Shanghai Aucy Technology Instrument Co., Ltd., Shanghai, China). RNA with A260/A280 values ranging from 1.8 to 2.0 was adjusted to a concentration of 50 ng/μL and used as a template for the RT reaction performed with a PrimeScriptTM RT Reagent Kit (RR047A, Takara, Beijing Think-far Technology Co., Ltd., Beijing, China). The RT reactions underwent 15 min reverse transcription at 37 °C and 5 s inactivation of reverse transcriptase at 85 °C. The total volume of the RT reaction system was 10 μL. The primers (shown in Table 1) were designed using Primer Premier 5.0 software (Premier Biosoft International, Palo Alto, CA) and synthesized by Tsingke Biological Technology Co., Ltd. (Beijing, China)18. Subsequently, RT-qPCR was performed on an ABI PRISM® 7900HT apparatus (ABI 7900, Shanghai PuDi Biotech Co., Ltd., Shanghai, China) using a two-step method. The RT-qPCR reaction conditions included pre-denaturation at 95 °C for 30 s, and 40 cycles of denaturation at 95 °C for 5 s, annealing at 58 °C for 30 s, and extension at 72 °C for 15 s. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were used as internal references for RT-qPCR. The relative mRNA expression of target genes was calculated using the 2−△△Ct method. Each target gene was tested in triplicate wells on the RT-qPCR plates for each sample.
For RT-qPCR, each experiment consisted of pre-denaturation at 95 °C for 30 s, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 58 °C for 30 s, and extension at 72 °C for 30 s. GAPDH was used as the internal reference for RT-PCR. Subsequently, electrophoresis was conducted, and each experiment was carried out 3 times.
Western blot analysis
Skeletal muscle tissues (30 mg in each group) were ground to a uniform powder at low temperature, washed 3 times with PBS, lysed with protein lysis buffer, incubated on ice for 20 min, and subsequently centrifuged at 12,000 r/min for 20 min. The supernatant was then collected, and protein concentration in the supernatant was determined using a bicinchoninic acid (BCA) protein assay kit (P0012-1; Beyotime Institute of Biotechnology Co., Shanghai, China). Cells in the logarithmic growth phase were centrifuged at 3000 r/min and 4 °C for 20 min to estimate the packed cell volume (PCV). Subsequently, 20 μL of PCV cells were lysed for 30 min with 100 μL of lysis buffer (containing 1 μL of enzyme inhibitor, 111111111, Roche; Beijing Jiamay Biotechnology Co., Ltd., Beijing, China) and centrifuged at 12,000 r/min and 4 °C for 10 min to collect the supernatant protein for Western blot analysis. The protein samples (50 µg in each group) were dissolved in 2× sodium dodecyl sulfate (SDS) sample buffer, boiled for 10 min, separated by 10% SDS-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred onto a polyvinylidene difluoride (PVDF) membrane. The membrane was then blocked in 5% skimmed milk at room temperature for 1 h, washed twice with PBS, and then incubated overnight at 4 °C with diluted rabbit anti-mouse primary antibodies against KCNMA1 (1:1000, MAB8589; AmyJet Scientific Inc., Wuhan, China), PKGII (1:1000, ab145063; Abcam, Inc., Cambridge, MA, USA), VASP (1:1000, ab58555; Abcam, USA), VEGF (1:1000, ab32152; Abcam, USA), Bax (1:1000, ab32503; Abcam, USA), Bcl-2 (1:1000, ab59348; Abcam, USA), Cyclin D1 (1:1000, ab134175, Abcam, USA), Cyclin D3 (1:1000, ab28283, Abcam, USA), Cdc 42 (1:1000, ab187643, Abcam, USA), caspase-3 (1:1000, ab208161, Abcam, USA), and PARP (1:1000, ab32064, Abcam, USA). After being washed 3 times in Tris-buffered saline tween-20 (TBST), the membrane was incubated with HRP-labeled goat anti-rabbit IgG secondary antibodies (1:5000) for 1 h and washed 3 times with TBST (5 min each time). An enhanced chemiluminescence (ECL) kit was then used for coloration. After liquid removal, the membrane was exposed to X-ray, and the absorbance of protein bands was analyzed using a gel imaging analysis system. In the analysis, GAPDH was used as the internal reference. Relative protein expression was calculated based on the ratio of the average absorbance of the target protein band to that of the internal reference band. Each experiment was conducted 3 times.
Cell treatment
Skeletal muscle tissues were randomly chosen from the normal and Sevo groups, rinsed and placed in clean and dry culture dishes. After being cut into small sections, the samples were sterilized with 0.01 M PBS (pH 7.6), centrifuged at 800 r/min for 5 min at room temperature, and digested with 0.5 mg/mL Type IV collagenase (17101-015; Gibco Company, Grand Island, NY, USA) at 37 °C for 15 min to collect the supernatant. The supernatant was then filtered with 40-μm cell mesh and centrifuged at 800 r/min for 5 min to collect the cells into a pellet. Subsequently, the cells were resuspended to a density of 2.5 × 107 cells/mL in RPMI 1640 (22400089; Gibco Company, Grand Island, NY, USA) medium containing 15% fetal bovine serum (FBS), seeded in a 6-well plate and cultured at 37 °C and 5% CO2 with saturate humidity. The culture medium was replaced every 2–3 days. After the single-layer cells reached approximately 80–90% confluence, they were passaged.
Cells in the logarithmic growth phase were divided into a control (skeletal muscle cells of normal mice), a blank group (skeletal muscle cells of mice in the Sevo group), a negative control group (NC; skeletal muscle cells of mice in the Sevo group and underwent no transfection), a MBNL1-AS1 vector group (skeletal muscle cells of Sevo mice transfected with MBNL1-AS1), a si-MBNL1-AS1#1 group (skeletal muscle cells of Sevo mice transfected with si-MBNL1-AS1), a si-MBNL1-AS1#2 group (skeletal muscle cells of Sevo mice transfected with si-MBNL1-AS1), a si-KCNMA1#1 group (skeletal muscle cells of Sevo mice transfected with si-KCNMA1), a si-KCNMA1#2 group (skeletal muscle cells of Sevo mice transfected with si-KCNMA1), and a si-MBNL1-AS1 + si-KCNMA1 group (skeletal muscle cells of Sevo mice transfected with si-MBNL1-AS1#1 and si-MBNL1-AS1#2 + si-KCNMA1#1 and si-KCNMA1#2) group. Lipofectamine (lipo) 2000 (Invitrogen, Inc., Carlsbad, CA, USA) was used for transfection per the manufacturer’s instructions. At 24–48 h after transfection, the cells were harvested.
Immunofluorescence assay
Cells were seeded onto a 96-well plate at a density of 1 × 105/mL and fixed in 4% polyoxymethylene for 15 min. After being washed thrice with PBS, the cells were treated with 0.5% TritonX 100 for 10 min and blocked with 5% goat serum at room temperature for 1 h. Subsequently, the cells were incubated with primary antibodies (ab9465 1: 500 αActinin, ab1835 1:500 TNNI1, and ab85087 1:50 Myogenin, Abcam, London, UK) at 4 °C overnight and then washed with PBS. Next, the cells were incubated with secondary antibodies at room temperature in the dark for 1 h. Finally, the cell nuclei were stained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) for 5 min, and the fluorescence signal in each sample was observed.
Dual-luciferase reporter gene assay
Putative target genes of lncRNA MBNL1-AS1 were determined using the program lncRNATargets (http://www.herbbol.org:8001/lrt/). Human embryonic kidney 293T (HEK-293T) cells were inoculated in a 24-well plate for 24 h and then co-transfected with luciferase reporter gene vectors KCNMA1 (pmiRRB- KCNMA1-3′UTR) + MBNL1-AS1 or siRNA-MBNL1-AS1, respectively. At 48 h after transfection, HEK-293FT cells were harvested, washed 2 times with PBS, and lysed. The luciferase reporter activity was detected using a Dual-Luciferase® Reporter Assay System (E1910; Promega, Madison, WI, USA). The relative luciferase reporter activity was calculated as the ratio of firefly luciferase activity to the luciferase activity of the Renilla internal control. The experiment was repeated 3 times.
RNA-immunoprecipitation (RIP)
Using PCR, the 5′ end of the MBNL1-AS1 gene was connected to a T7 promoter sequence (5′AGTAATACGACTCACTATAGGG3′). Subsequently, the products were extracted using a QIAquick Gel Extraction Kit (28704, Qiagen West Sussex, UK). Purified biotin-labeled MBNL1-AS1 RNA was transcribed in vitro with a MEGAshortscript™ T7 Transcription Kit (AM1354, Thermo Fisher Scientific, Waltham, MA, USA). Subsequently, we used a Pierce™ Magnetic RNA-Protein Pull-Down Kit (20164, Thermo Fisher Scientific) to pull down the proteins that interacted with RNA. Finally, the proteins were detected by Western blot analysis.
3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay
At 48 h after transfection, cells from each group were harvested and resuspended in Dulbecco’s minimum essential medium (DMEM) containing 10% FBS to a density of 1 × 105 cells/mL, which was then inoculated in a 96-well plate in 100 μL/well of medium. The cells were then cultured at 37 °C and under 5% CO2. At 24, 48, and 72 h of cell culture, 10 μL MTT (5 mg/mL; Sigma-Aldrich Chemical Company, St Louis, MO, USA) solution was added to the cells, which were further cultured for 4 h before the supernatant was discarded. Subsequently, 150 μL dimethyl sulphoxide (DMSO) was added to each well, and the optical density (OD) value of each well was measured by an automatic plate reader (Bio-Rad Laboratories, Hercules, CA, USA) at a wavelength of 490 nm. The experiment was repeated 3 times.
5-Ethynyl-2′-deoxyuridine (EdU) assay
Cells were seeded in triplicate onto a 96-well plate. On the following day, complete medium containing 50 µM EdU was added to the cells, which were further incubated at 5% CO2 and 37 °C for 2 h. After being washed thrice with PBS, the cells were fixed in 4% polyoxymethylene and then treated with 0.5% TritonX 100 for 5 min and incubated with an Apollo dye for 30 min. Subsequently, the cells were rinsed 3 times with 0.5% TritonX 100 and methanol and stained with DAPI for visualization and analysis. The experiment was repeated 3 times.
Flow cytometry
At 48 h after transfection, the cells were treated with 0.25% trypsin, and the cell density was adjusted to 1 × 106 cells/mL. Subsequently, 1 mL of cells was centrifuged at 1500 r/min for 10 min to collect the pellet, which was then fixed with 70% alcohol overnight at 4 °C. The next day, the cells were washed twice with PBS, and 100 μL of cell suspension was incubated in the dark for 30 min with 50 μg of RNAase-containing propidium iodide (PI, 40710ES03; Shanghai Qianchen Biological Technology Co., Ltd., Shanghai, China). Finally, after filtering the cells with a 100-mesh nylon net, the cell cycle in each sample was analyzed at a wavelength of 488 nm on a flow cytometer (BD Falcon, Franklin Lakes, NJ, USA).
Fluorescein isothiocyanate-labeled Annexin V/PI (Annexin V-FITC/PI) double-staining was used for the detection of cell apoptosis. In brief, after culturing the cells in a 5% CO2 incubator at 37 °C for 48 h, the cells were washed with PBS, centrifuged, and resuspended in 200 μL binding buffer. After adding 10 μL of Annexin V-FITC (ab14085; Abcam, Inc., MA, USA) and 5 μL of PI, the cells were incubated for 15 min at room temperature in the dark before 300 μL of binding buffer was added to each sample. Cell apoptosis was then analyzed at a wavelength of 488 nm on a flow cytometer (BD Falcon, Franklin Lakes, NJ, USA).
Statistical analysis
Data were analyzed using SPSS 21.0 software (IBM Corp., Armonk, NY, USA). Measurement data were expressed as the mean ± standard deviation. Comparisons between two groups were performed by t-test, while comparisons among multiple groups were performed by one-way analysis of variance (ANOVA). A p value of <0.05 was considered statistically significant.
Results
Identification of differentially expressed lncRNAs
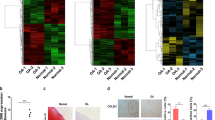
Initially, we used the GSE21164 microarray dataset from the GEO database to identify a differentially expressed lncRNA, MBNL1-AS1, which was overexpressed in I/R mice after TKA (Fig. 1).
KCNMA1 was a target gene of MBNL1-AS1
Subsequently, to verify the relationship between MBNL1-AS1 and KCNMA1, we conducted target gene prediction using MEM. As shown in Supplementary Table 1, KCNMA1 was found to be a target gene of MBNL1-AS1. Subsequently, KEGG enrichment analysis based on the WebGestalt database indicated that KCNMA1 was involved in the cGMP-PKG signaling pathway.
Histological changes in skeletal muscle tissues
In the next step, we observed histological changes in skeletal muscle tissues using HE staining, and the results are shown in Fig. 2. Skeletal muscle tissues in the normal group showed regularly arranged muscle fibers with clear bands, pink cytoplasm, and no signs of inflammatory cell infiltration, while the skeletal muscle tissues in the I/R group showed necrotic and disordered muscle fibers as well as signs of inflammatory cell infiltration, with a small amount of central nuclear fiber. The skeletal muscle tissues in the Sevo group mostly showed nicely arranged skeletal muscles with a small amount of inflammatory cell infiltration.
LDH, TNF-α, and IL-1β were increased in mice with I/R injury after TKA
We also used ELISA to measure the concentrations of LDH, TNF-α, and IL-1β in the plasma of the normal, I/R and Sevo groups. As shown in Table 2 and compared with the normal group, the concentrations of LDH, TNF-α, and IL-1β in the I/R and Sevo groups were significantly increased (p < 0.05). In comparison with the I/R group, the Sevo group showed decreased concentrations of LDH, TNF-α, and IL-1β (p < 0.05), suggesting that the concentrations of LDH, TNF-α, and IL-1β might be higher in mice with I/R injury after TKA.
Positive expression rate of KCNMA1 was down-regulated in mice with I/R injury after TKA
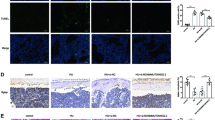
To clarify how KCNMA1 was expressed in mice with I/R injury after TKA, we performed immunohistochemistry assays. Results indicating positive expression of KCNMA1 in skeletal muscle tissues are shown in Fig. 3. KCNMA1 was mainly expressed in the cytoplasm, and the cells with brownish yellow cytoplasm were determined to be KCNMA1 positive. Compared with the normal group, the positive expression rate of KCNMA1 in the I/R and Sevo groups was markedly decreased (p < 0.05). In comparison with the I/R group, the Sevo group showed elevated KCNMA1 expression (p < 0.05), suggesting that KCNMA1 might be poorly expressed in I/R injury after TKA.
a Images of skeletal muscle tissues acquired after immunohistochemistry assays. b Positive expression rate of KCNMA1. I/R ischemia-reperfusion, Sevo sevoflurane, KCNMA1 potassium calcium-activated channel subfamily M alpha 1; *p < 0.05 vs. the normal group; # p < 0.05 vs. the I/R group; scale bar = 25 µm; n = 10; measurement data are expressed as the mean ± standard deviation. Comparisons among multiple groups were performed by one-way ANOVA; the experiment was repeated 3 times
High expression of MBNL1-AS1 and low expression of KCNMA1 and cGMP-PKG pathway-related factors in mice with I/R injury after TKA
To measure MBNL1-AS1, KCNMA1, and cGMP-PKG mRNA expression in mice with I/R injury after TKA, we performed RT-qPCR. As presented in Fig. 4a, KCNMA1, PKGII, VASP, VEGF, Bcl-2, Cyclin D1, Cyclin D3, and Cdc 42 mRNA expression was reduced in the I/R and Sevo groups compared with that in the normal group, but MBNL1-AS1 and Bax mRNA expression was elevated (p < 0.05). Compared with the I/R group, KCNMA1, PKGII, VASP, VEGF, Bcl-2, Cyclin D1, Cyclin D3, and Cdc 42 mRNA expression was increased in the Sevo group, while MBNL1-AS1 and Bax mRNA expression was decreased (p < 0.05).
a Relative mRNA expression of MBNL1-AS1, KCNMA1, PKGII, VASP, VEGF, Bax, and Bcl-2 in the skeletal muscle tissues of mice detected by RT-qPCR. b Protein bands observed after Western blot analysis. c Relative protein levels of KCNMA1, PKGII, VASP, p-PKGII, p-VASP, VEGF, Bax, and Bcl-2 in skeletal muscle tissues of mice evaluated by Western blot analysis. I/R ischemia-reperfusion, Sevo sevoflurane, MBNL1-AS1 muscleblind-like 1 antisense RNA 1, KCNMA1 potassium calcium-activated channel subfamily alpha 1, PKGII type II cyclic guanosine monophosphate-dependent protein kinase G, VASP vasodilator-stimulated phosphoprotein, VEGF vascular endothelial growth factor, Bcl-2 B-cell lymphoma/leukemia-2, Bax Bcl-2 associated X protein, GAPDH glyceraldehyde-3-phosphate dehydrogenase; *p < 0.05 vs. the normal group; # p < 0.05 vs. the I/R group; n = 10; measurement data are expressed as the mean ± standard deviation. Comparisons among multiple groups were performed by one-way ANOVA; the experiment was repeated 3 times
To measure the protein levels of MBNL1-AS1, KCNMA1 and cGMP-PKG in mice with I/R injury after TKA, we performed Western blot analyses. As shown in Fig. 4b, c, in comparison with the normal group, the protein levels of KCNMA1, PKGII, VASP, p-PKGII, p-VASP, VEGF, Bcl-2, Cyclin D1, Cyclin D3 and Cdc 42 were decreased in the I/R and Sevo groups, but the protein level of Bax was elevated (p < 0.05). The protein levels of KCNMA1, PKGII, VASP, p-PKGII, p-VASP, VEGF, Bcl-2, Cyclin D1, Cyclin D3 and Cdc 42 were increased in the Sevo group compared with those in the I/R group, but the protein levels of Bax, cleaved caspase-3, and cleaved PARP were reduced (p < 0.05). These results suggested that in the normal group, MBNL1-AS1 was poorly expressed but KCNMA1 was over-expressed and that the cGMP-PKG pathway was activated.
MBNL1-AS1 targeted and negatively regulated KCNMA1
Our previous prediction of target genes indicated that the mRNA of MBNL1-AS1 contained the binding site for KCNMA1 in its 3′ untranslated region (3′ UTR) (Fig. 5a). Compared with the NC group, the relative luciferase activity was decreased in the MBNL1-AS1 vector group but was elevated in the siRNA-MBNL1-AS1#1 and siRNA-MBNL1-AS1#2 groups (p < 0.05) (Fig. 5b). The results from the RIP assay showed no interaction between MBNL1-AS1 and KCNMA1 (Fig. 5c). These results suggested that MBNL1-AS1 might bind to KCNMA1 mRNA and inhibit its activity.
a The predicted consequential pairing between MBNL1-AS1 and KCNMA1. b Relative luciferase activity in the NC, MBNL1-AS1 vector, and siRNA-MBNL1-AS1 groups determined by the dual-luciferase reporter gene assay. c RIP results in each group. MBNL1-AS1 muscleblind-like 1 antisense RNA 1, KCNMA1 potassium calcium-activated channel subfamily M alpha 1, si small interfering, NC negative control; *p < 0.05 vs. the NC group; measurement data are expressed as the mean ± standard deviation. Comparisons among multiple groups were performed by one-way ANOVA; the experiment was repeated 3 times
Identification of skeletal muscle cells was confirmed
To confirm that the cells used in this experiment were skeletal muscle cells, we used RT-PCR and immunofluorescence tests. Figure 6 shows that αActinin, TNNI1, and Myogenin were positively expressed in these cells, suggesting that the cells used in this experiment were indeed skeletal muscle cells.
MBNL1-AS1 lowered the expression of KCNMA1 and cGMP-PKG pathway-related genes in skeletal muscle cells
In the subsequent experiment, we used RT-qPCR to assess the effects of MBNL1-AS1 on the mRNA expression of KCNMA1 and cGMP-PKG-related genes (shown in Fig. 7). Compared with the control group, KCNMA1, PKGII, VASP, VEGF, Bcl-2, Cyclin D1, Cyclin D3, and Cdc 42 mRNA expression was elevated in the blank, NC, MBNL1-AS1 vector, si-MBNL1-AS1#1, si-MBNL1-AS1#2, si-KCNMA1#1, si-KCNMA1#2, and si-MBNL1-AS1 + si-KCNMA1 groups, but MBNL1-AS1 and Bax mRNA expression was decreased (p < 0.05). In comparison with the blank and NC groups, MBNL1-AS1 mRNA expression was increased in the MBNL1-AS1 vector group, but Bax mRNA expression was elevated in the MBNL1-AS1 vector, si-KCNMA1#1 and si-KCNMA1#2 groups. In addition, KCNMA1, PKGII, VASP, VEGF, Bcl-2, Cyclin D1, Cyclin D3, and Cdc 42 mRNA expression was also reduced in the MBNL1-AS1 vector, si-KCNMA1#1 and si-KCNMA1#2 groups. Furthermore, KCNMA1, PKGII, VASP, VEGF, Bcl-2, Cyclin D1, Cyclin D3, and Cdc 42 mRNA expression was increased in the si-MBNL1-AS1#1 and si-MBNL1-AS1#2 groups, but the expression of MBNL1-AS1 and Bax was decreased (p < 0.05). In the si-MBNL1-AS1 + si-KCNMA1 group, only MBNL1-AS1 mRNA expression was decreased (p > 0.05). (Because the results of groups #1 and #2 were similar, only the results of one group were shown.)
a RT-qPCR measured the relative mRNA expression of MBNL1-AS1, KCNMA1, PKGII, VASP, VEGF, Bax, and Bcl-2 in skeletal muscle cells after transfection. b Protein bands measured by Western blot analysis. c Western blot analysis measured the relative protein levels of KCNMA1, PKGII, VASP, p-PKGII, p-VASP, VEGF, Bax, and Bcl-2 in skeletal muscle cells after transfection. MBNL1-AS1 muscleblind-like 1 antisense RNA 1, KCNMA1 potassium calcium-activated channel subfamily M alpha 1, PKGII type II cyclic guanosine monophosphate-dependent protein kinase G, p-PKGII phosphorylated-PKGII, VASP vasodilator-stimulated phosphoprotein, p-VASP phosphorylated-VASP, VEGF vascular endothelial growth factor, Bcl-2 B-cell lymphoma/leukemia-2, Bax Bcl-2 associated X protein, si small interfering, GAPDH glyceraldehyde-3-phosphate dehydrogenase, NC negative control; *p < 0.05 vs. the control group; # p < 0.05 vs. the blank and NC groups; measurement data are expressed as the mean ± standard deviation. Comparisons among multiple groups were done. Comparisons among multiple groups were performed by one-way ANOVA; the experiment was repeated 3 times
Using Western blot analysis, we also assessed the effects of MBNL1-AS1 on the mRNA expression of KCNMA1 and cGMP-PKG-related genes (shown in Fig. 7). In comparison with those in the control group, the protein levels of KCNMA1, PKGII, VASP, p-PKGII, p-VASP, VEGF, Bcl-2, Cyclin D1, Cyclin D3, and Cdc 42 were increased in the blank, NC, MBNL1-AS1 vector, si-MBNL1-AS1#1, si-MBNL1-AS1#2, si-KCNMA1#1, si-KCNMA1#2, and si-MBNL1-AS1 + si-KCNMA1 groups, but the protein levels of Bax, cleaved caspase-3, and cleaved PARP were reduced in these groups (p < 0.05). Compared with those in the blank and NC groups, the protein levels of KCNMA1, PKGII, VASP, p-PKGII, p-VASP, VEGF, Bcl-2, Cyclin D1, Cyclin D3, and Cdc 42 were decreased in the MBNL1-AS1 vector, si-KCNMA1#1, and si-KCNMA1#2 groups, but the protein levels of Bax, cleaved caspase-3, and cleaved PARP were increased in these groups. While the protein levels of KCNMA1, PKGII, VASP, p-PKGII, p-VASP, VEGF, Bcl-2, Cyclin D1, Cyclin D3, Cdc 42, cleaved caspase-3, and cleaved PARP were all elevated in the si-MBNL1-AS1#1 and si-MBNL1-AS1#2 groups, the protein levels of Bax, cleaved caspase-3, and cleaved PARP (p < 0.05) were decreased in these groups. These results indicated that the KCNMA1 and cGMP-PKG pathways might be negatively regulated by MBNL1-AS1, while the overexpression of KCNMA1 could increase the expression of cGMP-PKG pathway-related proteins suppressed by the overexpression of MBNL1-AS1.
MBNL1-AS1 inhibited the proliferation of skeletal muscle cells
Using MTT and EdU assays, we investigated the effects of MBNL1-AS1 on skeletal muscle cell proliferation. As shown in Fig. 8, the OD values of skeletal muscle cells among the seven groups started to show significant differences after 48 h of cell culture. Compared with cells in the control group (p < 0.05), cells in the blank, NC, MBNL1-AS1 vector, si-MBNL1-AS1#1, si-MBNL1-AS1#2, si-KCNMA1#1, si-KCNMA1#2, and si-MBNL1-AS1 + si-KCNMA1 groups began to grow faster after 48 h. In comparison with cells in the blank and NC groups, cells in the MBNL1-AS1 vector and si-KCNMA1 groups grew slower but grew faster in the si-MBNL1-AS1#1 and si-MBNL1-AS1#2 groups (p < 0.05), while no significant differences were shown in the si-MBNL1-AS1 + si-KCNMA1 group (p > 0.05). The results of the EdU assay were consistent with those of the MTT assay. In comparison with the blank and NC groups, the cells in the MBNL1-AS1 vector, si-KCNMA1#1, and si-KCNMA1#2 groups grew slower, but the cells in the si-MBNL1-AS1#1 and si-MBNL1-AS1#2 groups (p < 0.05) grew faster, while no obvious differences were found in the si-MBNL1-AS1 + si-KCNMA1 group (p > 0.05). (Because the results of #1 and #2 were similar, only the results from one group are shown.) These results showed that MBNL1-AS1 could inhibit the proliferation of skeletal muscle cells, while KCNMA1 could promote cell growth. In addition, the overexpression of KCNMA1 alleviated the inhibitory effect of MBNL1-AS1 on the proliferation of skeletal muscle cells.
a Cell proliferation in each group as measured by MTT. b Cell proliferation in each group as measured by EdU. c Cell proliferation in each group as measured by EdU (×50). OD optical density, MBNL1-AS1 muscleblind-like 1 antisense RNA 1, KCNMA1 potassium calcium-activated channel subfamily M alpha 1, si small interfering, NC negative control; *p < 0.05 vs. the control group; # p < 0.05 vs. the blank and NC groups; scale bar = 200 µm; the experiment was repeated 3 times
MBNL1-AS1 promoted the apoptosis of skeletal muscle cells
PI staining was performed to evaluate the effects of MBNL1-AS1 on cell cycle distribution (Fig. 9a). Compared with the control group, the number of cells in G0/G1 phase was decreased in the blank, NC, MBNL1-AS1 vector, si-MBNL1-AS1#1, si-MBNL1-AS1#2, si-KCNMA1#1, si-KCNMA1#2, and si-MBNL1-AS1 + si-KCNMA1 groups, but the number of cells in S phase was increased in these groups (p < 0.05). In comparison with the blank and NC groups, the number of cells in G0/G1 phase was increased in the MBNL1-AS1 vector, si-KCNMA1#1, and si-KCNMA1#2 groups, but the number of cells in S phase was decreased in these groups. In addition, the number of cells in G0/G1 phase was decreased in the si-MBNL1-AS1#1 and si-MBNL1-AS1#2 groups, while the number of cells in S phase was increased in these groups (p < 0.05). Furthermore, no significant differences were shown in the si-MBNL1-AS1 + si-KCNMA1 group (p > 0.05). Annexin V-FITC/PI staining was conducted to evaluate the effects of MBNL1-AS1 on cell apoptosis (Fig. 9b). Compared with that in the control group, the apoptosis rate of skeletal muscle cells was reduced in the other six groups. In comparison with that in the blank and NC groups, the cell apoptosis rate was elevated in the MBNL1-AS1 vector, si-KCNMA1#1, and si-MBNL1-AS1#2 groups (p < 0.05), while the apoptosis rate was decreased in the si-MBNL1-AS1#1 and si-MBNL1-AS1#2 groups (p < 0.05). In addition, no significant differences were observed in the si-MBNL1-AS1 + si-KCNMA1 group. These results suggested that MBNL1-AS1 could promote the apoptosis of skeletal muscle cells, while KCNMA1 could inhibit the apoptosis of these cells. In addition, the overexpression of KCNMA1 alleviated the effect caused by MBNL1-AS1 overexpression (Table 3).
a Cell cycle of skeletal muscle cells in seven groups as detected by flow cytometry. b Apoptosis of skeletal muscle cells in seven groups as detected by flow cytometry. MBNL1-AS1 muscleblind-like 1 antisense RNA 1, KCNMA1 potassium calcium-activated channel subfamily M alpha 1, si small interfering, NC negative control; *p < 0.05 vs. the control group; # p < 0.05 vs. the blank and NC groups; measurement data are expressed as the mean ± standard deviation. Comparisons among multiple groups were performed by one-way ANOVA; the experiment was repeated 3 times
Discussion
As the most common degenerative joint disorder, OA is a major cause of pain and disability worldwide, and the medical therapy of its complications remains ineffective due to poor understanding of its pathogenesis19. In end-stage knee OA, TKA might be the best choice for treatment, but it also places patients at serious risk of I/R injury20,21. In our experiment, a mouse model of I/R injury after TKA was successfully established, and the collective results demonstrated that down-regulated MBNL1-AS1 might increase the expression of KCNMA1 through activation of the cGMP-PKG signaling pathway, which in turn resulted in the promotion of skeletal muscle cell proliferation and the suppression of their apoptosis. Therefore, down-regulated MBNL1-AS1 might protect Sevo-pretreated mice against I/R injury after TKA.
Sevo has achieved initial success in protecting against I/R injury6,7. Unfortunately, several studies have indicated that the inhalation of Sevo might cause apoptosis in the brain tissues of neonatal mice22,23. lncRNA, a noncoding RNA of more than 200 nucleotides, can regulate tissue homeostasis and pathophysiological conditions by interfering with gene expression, although the molecular mechanisms of lncRNAs in human disorders are still poorly understood24,25. Our results indicated that lncRNA MBNL1-AS1 was overexpressed in the skeletal muscle cells of mice suffering from I/R injury after TKA. In fact, some scientists have stressed the advantages of using lncRNAs to improve the function of skeletal muscles26. Thus, lncRNA MBNL1-AS1 might become an important target for the rehabilitation of I/R injury after TKA, although further studies are required to understand its underlying mechanisms.
Represented by immune and inflammatory responses, the patterns of lncRNA–mRNA co-expression have also been shown in muscle contraction27. In our study, the co-expression patterns of lncRNA MBNL1-AS1 and KCNMA1 in the metabolism of skeletal muscle cells were also studied. We proved that KCNMA1 was negatively regulated by MBNL1-AS1, indicating that the role of MBNL1-AS1 in the protection of skeletal muscle cells against I/R injury might be associated with KCNMA1. As a type of gene encoding pore-forming potassium big-conductance calcium-activated (BK) channel proteins in the cell membrane, KCNMA1 mainly acts to maintain the intracellular and extracellular balance of K+ and Ca2+ , as well as to stabilize membrane potential and regulate the contraction of vascular smooth muscle cells28. Moreover, a previous study demonstrated that KCNMA1 showed a positive effect on the survival of cardiomyocytes after I/R injury, while cardiac BK channels encoded by KCNMA1 could protect against I/R injury through the participation of reactive oxygen species (ROS)29. In addition, the involvement of KCNMA1 in the modulation of excitability in skeletal muscles might explain the oxidative muscle injury and muscle atrophy caused by I/R injury after TKA30. In addition, KEGG enrichment analyses indicated that KCNMA1 was enriched in the cGMP-PKG signaling pathway, which could regulate diverse cellular functions and gene expression patterns31,32,33,34. Such results were in line with a previous discovery showing that testosterone-induced relaxation of porcine coronary arteries relied on the activation of BK channels via the cGMP-PKG signaling pathway35. In summary, we speculated that the expression of KCNMA1 increased by the down-regulation of MBNL1-AS1 opened the BK channels via the cGMP-PKG signaling pathway to protect skeletal muscle cells against I/R injury after TKA.
LDH not only promotes the interconversion of pyruvate and lactate but also serves as a key metabolite of skeletal muscle and promotes muscle mitochondrial functions36. Further studies also demonstrated that extra-matrix LDH is strategically positioned within skeletal muscle fibers to functionally interact with mitochondria and that the suppressed mitochondrial pyruvate supply can inhibit mitochondrial respiration37. A previous study also found that LDH activity is elevated in the plasma during I/R injury, consistent with our results38. In the I/R group, skeletal muscle tissues showed necrotic muscle fibers and inflammatory cell infiltration, signs indicative of serious nuclear deformation and fibrinolysis. Moreover, it is well known that myocytes preconditioned by exposure to a PKG activator (Sp-8-Br-PET-cGMPS) show a marked decrease in DNA and LDH damage, either due to the direct activation of ATP-sensitive K+ channels or via the PKG signaling pathway, thus leading to cardioprotection39. The increased levels of TNF-α and IL-1β, two circulatory markers of inflammation, in the serum at the early phase of reperfusion can further lead to more severe I/R injury through inflammatory response and oxidative stress40,41.
In conclusion, our data may explain the mechanism underlying the effect of the down-regulation of MBNL1-AS1 and the subsequent promotion of the proliferation and inhibition of skeletal muscle cell apoptosis, as we found that the down-regulation of MBNL1-AS1 exerted its effect by up-regulating the expression of KCNMA1 and by activating the cGMP-PKG signaling pathway. As a result, the down-regulation of MBNL1-AS1 protected Sevo-pretreated mice against I/R injury after TKA. Although the factors and signaling pathways that regulate the metabolism of skeletal muscle cells are complicated and still poorly understood, we still believe our findings may provide a rational basis for the clinical application of MBNL1-AS1 as a target in the treatment of I/R injury after TKA.
Change history
08 November 2021
This article has been retracted. Please see the Retraction Notice for more detail: https://doi.org/10.1038/s12276-021-00698-5
References
Chiang, C. Y., Chen, K. H., Liu, K. C., Hsu, S. J. & Chan, C. T. Data collection and analysis using wearable sensors for monitoring knee range of motion after total knee arthroplasty. Sensors (Basel) 17, 418 (2017).
Xu, Z. H. et al. [Preliminary clinical outcome of domestic posterior-stabilized total knee arthroplasty]. Zhongguo Gu Shang 22, 410–412 (2009).
Shadgan, B. et al. Hemodynamic and oxidative mechanisms of tourniquet-induced muscle injury: near-infrared spectroscopy for the orthopedics setting. J. Biomed. Opt. 17, 081408 (2012).
Kocman, E. A. et al. Effects of ischemic preconditioning protocols on skeletal muscle ischemia-reperfusion injury. J. Surg. Res. 193, 942–952 (2015).
Muyskens, J. B. et al. Transcriptional profiling and muscle cross-section analysis reveal signs of ischemia reperfusion injury following total knee arthroplasty with tourniquet. Physiol. Rep. 4, e12671 (2016).
Wu, Y., Gu, C. & Huang, X. Sevoflurane protects against hepatic ischemia/reperfusion injury by modulating microRNA-200c regulation in mice. Biomed. Pharmacother. 84, 1126–1136 (2016).
Cao, J. et al. Sevoflurane post-conditioning reduces rat myocardial ischemia reperfusion injury through an increase in NOS and a decrease in phopshorylated NHE1 levels. Int. J. Mol. Med. 36, 1529–1537 (2015).
Jones, M. R. et al. Systems genetics reveals the functional context of PCOS loci and identifies genetic and molecular mechanisms of disease heterogeneity. PLoS Genet. 11, e1005455 (2015).
Sha, Y. et al. Protective effect of ischaemic preconditioning in total knee arthroplasty. Eur. Rev. Med. Pharmacol. Sci. 18, 1559–1566 (2014).
Carithers, L. J. & Moore, H. M. The Genotype-Tissue Expression (GTEx) project. Biopreserv. Biobank. 13, 307–308 (2015).
Adler, P. et al. Mining for coexpression across hundreds of datasets using novel rank aggregation and visualization methods. Genome Biol. 10, R139 (2009).
Zhang, J. et al. Gene ontology and KEGG enrichment analyses of genes related to age-related macular degeneration. Biomed. Res. Int. 2014, 450386 (2014).
Wang, J., Vasaikar, S., Shi, Z., Greer, M. & Zhang, B. WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 45(W1), W130–W137 (2017).
Zhan, R. et al. Nitric oxide enhances keratinocyte cell migration by regulating Rho GTPase via cGMP-PKG signalling. PLoS One 10, e0121551 (2015).
Fujita, A., Sato, J. R., de Oliveira Rodrigues, L., Ferreira, C. E. & Sogayar, M. C. Evaluating different methods of microarray data normalization. BMC Bioinformatics 7, 469 (2006).
Smyth, G. K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, 3 (2004).
Wang, J., Duncan, D., Shi, Z. & Zhang, B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 41, W77–W83 (2013).
Li, X. et al. Overexpression of miR-708 and its targets in the childhood common precursor B-cell ALL. Pediatr. Blood Cancer 60, 2060–2067 (2013).
Xie, L. et al. Systemic neutralization of TGF-beta attenuates osteoarthritis. Ann. N. Y. Acad. Sci. 1376, 53–64 (2016).
van der Woude, J. A. et al. Knee joint distraction compared to total knee arthroplasty for treatment of end stage osteoarthritis: simulating long-term outcomes and cost-effectiveness. PLoS ONE 11, e015524 (2016).
Ejaz, A. et al. Tourniquet induced ischemia and changes in metabolism during TKA: a randomized study using microdialysis. BMC Musculoskelet. Disord. 16, 326 (2015).
Lu, Y. et al. Anesthetic sevoflurane causes neurotoxicity differently in neonatal naive and Alzheimer disease transgenic mice. Anesthesiology 112, 1404–1416 (2010).
Wang, M., Zhang, J. H. & Applegate, R. L. 2nd Adverse effect of inhalational anesthetics on the developing brain. Med. Gas Res. 4, 2 (2014).
Uchida, S. & Dimmeler, S. Long noncoding RNAs in cardiovascular diseases. Circ. Res. 116, 737–750 (2015).
Yao, Q. et al. Global prioritizing disease candidate lncRNAs via a multi-level composite network. Sci. Rep. 7, 39516 (2017).
Simionescu-Bankston, A. & Kumar, A. Noncoding RNAs in the regulation of skeletal muscle biology in health and disease. J. Mol. Med. 94, 853–866 (2016).
Wang, W. et al. Aberrant expression of lncRNAs and mRNAs in patients with intracranial aneurysm. Oncotarget 8, 2477–2484 (2017).
He, Y., He, W., Qin, G., Luo, J. & Xiao, M. Transplantation KCNMA1 modified bone marrow-mesenchymal stem cell therapy for diabetes mellitus-induced erectile dysfunction. Andrologia 46, 479–486 (2014).
Soltysinska, E. et al. KCNMA1 encoded cardiac BK channels afford protection against ischemia-reperfusion injury. PLoS ONE 9, e103402 (2014).
Gambara, G. et al. Gene expression profiling in slow-type calf soleus muscle of 30 days space-flown mice. PLoS ONE 12, e0169314 (2017).
Tian, W. et al. Identification of miRNAs and differentially expressed genes in early phase non-small cell lung cancer. Oncol. Rep. 35, 2171–2176 (2016).
Greenwood, C. et al. Stat1 and CD74 overexpression is co-dependent and linked to increased invasion and lymph node metastasis in triple-negative breast cancer. J. Proteomics 75, 3031–3040 (2012).
Takimoto, E. Cyclic GMP-dependent signaling in cardiac myocytes. Circ. J. 76, 1819–1825 (2012).
Ping, J. & Schafe, G. E. The NO-cGMP-PKG signaling pathway coordinately regulates ERK and ERK-driven gene expression at pre- and postsynaptic sites following LTP-inducing stimulation of thalamo-amygdala synapses. Neural Plast. 2010, 540940 (2010).
Deenadayalu, V., Puttabyatappa, Y., Liu, A. T., Stallone, J. N. & White, R. E. Testosterone-induced relaxation of coronary arteries: activation of BKCa channels via the cGMP-dependent protein kinase. Am. J. Physiol. Heart Circ. Physiol. 302, H115–H123 (2012).
Liang, X. et al. Exercise inducible lactate dehydrogenase B regulates mitochondrial function in skeletal muscle. J. Biol. Chem. 291, 25306–25318 (2016).
Elustondo, P. A. et al. Physical and functional association of lactate dehydrogenase (LDH) with skeletal muscle mitochondria. J. Biol. Chem. 288, 25309–25317 (2013).
Nielsen, P. M. et al. In situ lactate dehydrogenase activity: a novel renal cortical imaging biomarker of tubular injury? Am. J. Physiol. Ren. Physiol. 312, F465–F473 (2017).
Cuong, D. V. et al. Nitric oxide-cGMP-protein kinase G signaling pathway induces anoxic preconditioning through activation of ATP-sensitive K+ channels in rat hearts. Am. J. Physiol. Heart Circ. Physiol. 290, H1808–H1817 (2006).
Zhao, X. Q., Liang, B., Liu, Y. & Huang, X. Q. Agaricoglycerides protect against hepatic ischemia/reperfusion injury by attenuating inflammatory response, oxidative stress, and expression of NF-kappaB. Evid. Based Complement. Alternat. Med. 2015, 142736 (2015).
Klune, J. R. & Tsung, A. Molecular biology of liver ischemia/reperfusion injury: established mechanisms and recent advancements. Surg. Clin. North Am. 90, 665–677 (2010).
Acknowledgements
This study was supported by the National Youth Science Foundation (No. 80151459) and the Education Department of Jilin Province 13th Five-Year Science and Technology Research Project (2016-No. 467). We thank the people who provided assistance and helpful discussion for their contributions to our manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, XF., Wang, ZQ., Li, LY. et al. RETRACTED ARTICLE: Downregulation of the long noncoding RNA MBNL1-AS1 protects sevoflurane-pretreated mice against ischemia-reperfusion injury by targeting KCNMA1. Exp Mol Med 50, 1–16 (2018). https://doi.org/10.1038/s12276-018-0133-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s12276-018-0133-y
This article is cited by
-
microRNA-301b-3p downregulation underlies a novel inhibitory role of long non-coding RNA MBNL1-AS1 in non-small cell lung cancer
Stem Cell Research & Therapy (2019)