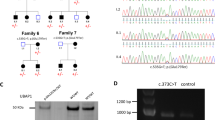
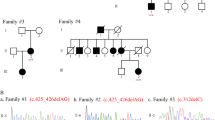
Abstract
Hereditary spastic paraplegias (HSPs) are a heterogeneous group of neurodegenerative disorders characterized by progressive spasticity and weakness in the lower extremities. To date, a total of 88 types of SPG are known. To diagnose HSP, multiple technologies, including microarray, direct sequencing, multiplex ligation-dependent probe amplification, and short-read next-generation sequencing, are often chosen based on the frequency of HSP subtypes. Exome sequencing (ES) is commonly used. We used ES to analyze ten cases of HSP from eight families. We identified pathogenic variants in three cases (from three different families); however, we were unable to determine the cause of the other seven cases using ES. We therefore applied long-read sequencing to the seven undetermined HSP cases (from five families). We detected intragenic deletions within the SPAST gene in four families, and a deletion within PSEN1 in the remaining family. The size of the deletion ranged from 4.7 to 12.5 kb and involved 1–7 exons. All deletions were entirely included in one long read. We retrospectively performed an ES-based copy number variation analysis focusing on pathogenic deletions, but were not able to accurately detect these deletions. This study demonstrated the efficiency of long-read sequencing in detecting intragenic pathogenic deletions in ES-negative HSP patients.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Saputra L, Kumar KR. Challenges and controversies in the genetic diagnosis of hereditary spastic paraplegia. Curr Neurol Neurosci Rep. 2021;21:15.
Meyyazhagan A, Orlacchio A. Hereditary spastic paraplegia: an update. Int J Mol Sci. 2022;23:1697.
Schob C, Hempel M, Safka Brozkova D, Jiang H, Kim SY, Batzir NA, et al. Dominant KPNA3 mutations cause infantile-onset hereditary spastic paraplegia. Ann Neurol. 2021;90:738–50.
Koh K, Ishiura H, Tsuji S, Takiyama Y. JASPAC: Japan Spastic Paraplegia Research Consortium. Brain Sci. 2018;8:153.
Shribman S, Reid E, Crosby AH, Houlden H, Warner TT. Hereditary spastic paraplegia: from diagnosis to emerging therapeutic approaches. Lancet Neurol. 2019;18:1136–46.
Hensiek A, Kirker S, Reid E. Diagnosis, investigation and management of hereditary spastic paraplegias in the era of next-generation sequencing. J Neurol. 2015;262:1601–12.
Gilissen C, Hoischen A, Brunner HG, Veltman JA. Unlocking Mendelian disease using exome sequencing. Genome Biol. 2011;12:228.
Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12:745–55.
Fromer M, Purcell SM. Using XHMM software to detect copy number variation in whole-exome sequencing data. Curr Protoc Hum Genet. 2014;81:7.23.1–21.
Nord AS, Lee M, King MC, Walsh T. Accurate and exact CNV identification from targeted high-throughput sequence data. BMC Genomics. 2011;12:184.
Uchiyama Y, Yamaguchi D, Iwama K, Miyatake S, Hamanaka K, Tsuchida N, et al. Efficient detection of copy-number variations using exome data: batch- and sex-based analyses. Hum Mutat. 2021;42:50–65.
Mantere T, Kersten S, Hoischen A. Long-read sequencing emerging in medical genetics. Front Genet. 2019;10:426.
Treangen TJ, Salzberg SL. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat Rev Genet. 2011;13:36–46.
Huddleston J, Chaisson MJP, Steinberg KM, Warren W, Hoekzema K, Gordon D, et al. Discovery and genotyping of structural variation from long-read haploid genome sequence data. Genome Res. 2017;27:677–85.
Benjamini Y, Speed TP. Summarizing and correcting the GC content bias in high-throughput sequencing. Nucleic Acids Res. 2012;40:e72.
van Dijk EL, Jaszczyszyn Y, Naquin D, Thermes C. The third revolution in sequencing technology. Trends Genet. 2018;34:666–81.
Mizuguchi T, Suzuki T, Abe C, Umemura A, Tokunaga K, Kawai Y, et al. A 12-kb structural variation in progressive myoclonic epilepsy was newly identified by long-read whole-genome sequencing. J Hum Genet. 2019;64:359–68.
Seyama R, Tsuchida N, Okada Y, Sakata S, Hamada K, Azuma Y, et al. Two families with TET3-related disorder showing neurodevelopmental delay with craniofacial dysmorphisms. J Hum Genet. 2022;67:157–64.
Danis D, Jacobsen JOB, Balachandran P, Zhu Q, Yilmaz F, Reese J, et al. SvAnna: efficient and accurate pathogenicity prediction of coding and regulatory structural variants in long-read genome sequencing. Genome Med. 2022;14:44.
Mitsuhashi S, Ohori S, Katoh K, Frith MC, Matsumoto N. A pipeline for complete characterization of complex germline rearrangements from long DNA reads. Genome Med. 2020;12:67.
MacDonald JR, Ziman R, Yuen RK, Feuk L, Scherer SW. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 2014;42:D986–92.
Ishiura H, Takahashi Y, Hayashi T, Saito K, Furuya H, Watanabe M, et al. Molecular epidemiology and clinical spectrum of hereditary spastic paraplegia in the Japanese population based on comprehensive mutational analyses. J Hum Genet. 2014;59:163–72.
Schüle R, Wiethoff S, Martus P, Karle KN, Otto S, Klebe S, et al. Hereditary spastic paraplegia: clinicogenetic lessons from 608 patients. Ann Neurol. 2016;79:646–58.
Solowska JM, Baas PW. Hereditary spastic paraplegia SPG4: what is known and not known about the disease. Brain 2015;138:2471–84.
Chelban V, Breza M, Szaruga M, Vandrovcova J, Murphy D, Lee CJ, et al. Spastic paraplegia preceding PSEN1-related familial Alzheimer’s disease. Alzheimers Dement. 2021;13:e12186.
Boone PM, Yuan B, Campbell IM, Scull JC, Withers MA, Baggett BC, et al. The Alu-rich genomic architecture of SPAST predisposes to diverse and functionally distinct disease-associated CNV alleles. Am J Hum Genet. 2014;95:143–61.
Solowska JM, D’Rozario M, Jean DC, Davidson MW, Marenda DR, Baas PW. Pathogenic mutation of spastin has gain-of-function effects on microtubule dynamics. J Neurosci. 2014;34:1856–67.
Solowska JM, Rao AN, Baas PW. Truncating mutations of SPAST associated with hereditary spastic paraplegia indicate greater accumulation and toxicity of the M1 isoform of spastin. Mol Biol Cell. 2017;28:1728–37.
Qiang L, Piermarini E, Baas PW. New hypothesis for the etiology of SPAST-based hereditary spastic paraplegia. Cytoskeleton. 2019;76:289–97.
Chen R, Du S, Yao Y, Zhang L, Luo J, Shen Y, et al. A novel SPAST mutation results in spastin accumulation and defects in microtubule dynamics. Mov Disord. 2022;37:598–607.
Solowska JM, Morfini G, Falnikar A, Himes BT, Brady ST, Huang D, et al. Quantitative and functional analyses of spastin in the nervous system: implications for hereditary spastic paraplegia. J Neurosci. 2008;28:2147–57.
Bagaria J, Bagyinszky E, An SSA. Genetics, functions, and clinical impact of presenilin-1 (PSEN1) gene. Int J Mol Sci. 2022;23:10970.
Hernandez-Sapiens MA, Reza-Zaldívar EE, Márquez-Aguirre AL, Gómez-Pinedo U, Matias-Guiu J, Cevallos RR, et al. Presenilin mutations and their impact on neuronal differentiation in Alzheimer’s disease. Neural Regen Res. 2022;17:31–7.
Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–6.
De Strooper B. Loss-of-function presenilin mutations in Alzheimer disease. Talking Point on the role of presenilin mutations in Alzheimer disease. EMBO Rep. 2007;8:141–6.
Watanabe H, Shen J. Dominant negative mechanism of Presenilin-1 mutations in FAD. Proc Natl Acad Sci USA. 2017;114:12635–7.
Kelleher RJ 3rd, Shen J. Presenilin-1 mutations and Alzheimer’s disease. Proc Natl Acad Sci USA. 2017;114:629–31.
Hardy J. The amyloid hypothesis for Alzheimer’s disease: a critical reappraisal. J Neurochem. 2009;110:1129–34.
Shen J, Kelleher RJ 3rd. The presenilin hypothesis of Alzheimer’s disease: evidence for a loss-of-function pathogenic mechanism. Proc Natl Acad Sci USA. 2007;104:403–9.
Heilig EA, Gutti U, Tai T, Shen J, Kelleher RJ 3rd. Trans-dominant negative effects of pathogenic PSEN1 mutations on γ-secretase activity and Aβ production. J Neurosci. 2013;33:11606–17.
Crook R, Verkkoniemi A, Perez-Tur J, Mehta N, Baker M, Houlden H, et al. A variant of Alzheimer’s disease with spastic paraparesis and unusual plaques due to deletion of exon 9 of presenilin 1. Nat Med. 1998;4:452–5.
Woodruff G, Young JE, Martinez FJ, Buen F, Gore A, Kinaga J, et al. The presenilin-1 ΔE9 mutation results in reduced γ-secretase activity, but not total loss of PS1 function, in isogenic human stem cells. Cell Rep. 2013;5:974–85.
Tsuchida N, Nakashima M, Kato M, Heyman E, Inui T, Haginoya K, et al. Detection of copy number variations in epilepsy using exome data. Clin Genet. 2018;93:577–87.
Miyatake S, Koshimizu E, Fujita A, Fukai R, Imagawa E, Ohba C, et al. Detecting copy-number variations in whole-exome sequencing data using the eXome Hidden Markov Model: an ‘exome-first’ approach. J Hum Genet. 2015;60:175–82.
Yao R, Zhang C, Yu T, Li N, Hu X, Wang X, et al. Evaluation of three read-depth based CNV detection tools using whole-exome sequencing data. Mol Cytogenet. 2017;10:30.
Acknowledgements
We would like to thank all the subjects for participating in this study. We also thank N. Watanabe, T. Miyama, M. Sato, S. Sugimoto, and K. Takabe for technical assistance. We are grateful to Anahid Pinchis and Alison Inglis, PhD, from Edanz (https://jp.edanz.com/ac) for editing a draft of this manuscript. This work was supported by the Japan Agency for Medical Research and Development (AMED) under grant numbers JP22ek0109595h0001 (HD) and JP22ek0109486, JP22ek0109549, and JP22ek0109493 (NM); JSPS KAKENHI under grant numbers JP23H02829 (SM), JP21K15907 (YU), JP22K15646 (KH), JP21K07869 (EK), JP22K15901 (AF), JP21K07298 (HD), JP21K07440 (FT) and JP23H02877 (TM); the Takeda Science Foundation (HD, NM, and TM); Health Labour Sciences Research Grants from the Ministry of Health, Labour and Welfare (202011073A, 202011029A; FT) and a grant for Strategic Research Promotion from Yokohama City University (SK2804; FT).
Author information
Authors and Affiliations
Contributions
HF and NM contributed to the study design. HF, HD, SK, SO, EK, YU, NT, AF, KH, KM, SM, TM and NM performed the data analysis. HD, MK, HJ, TT, HK, MS, YM and FT evaluated clinical information of patients. HF, TM and NM prepared the manuscript. NM finalized the manuscript. All authors critically reviewed and revised the manuscript and approved the final version for submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Fukuda, H., Mizuguchi, T., Doi, H. et al. Long-read sequencing revealing intragenic deletions in exome-negative spastic paraplegias. J Hum Genet 68, 689–697 (2023). https://doi.org/10.1038/s10038-023-01170-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-023-01170-0