Abstract
We aimed to find a new causative gene and elucidate the molecular mechanisms underlying a new type of hereditary spastic paraplegia (HSP). Patients with HSP were recruited from the Japan Spastic Paraplegia Research Consortium (JASPAC). Exome sequencing of genomic DNA from patients in four families was carried out, followed by Sanger sequencing of the UBAP1 gene. A mouse homolog of one UBAP1 frameshift mutation carried by one of the patients was created as a disease model. Functional properties of the UBAP1 wild type and UBAP1-mutant in mouse hippocampus neurons were examined. We identified three novel heterozygous loss of function mutations (c.425_426delAG, c.312delC, and c.535G>T) in the UBAP1 gene as the genetic cause of a new type of HSP (SPG80). All the patients presented identical clinical features of a pure type of juvenile-onset HSP. Functional studies on mouse hippocampal neurons revealed that the C-terminal deletion UBAP1-mutant of our disease model had lost its ability to bind ubiquitin in vitro. Overexpression of the UBAP1 wild type interacts directly with ubiquitin on enlarged endosomes, while the UBAP1-mutant cannot be recruited to endosome membranes. Our study demonstrated that mutations in the UBAP1 gene cause a new type of HSP and elucidated its pathogenesis. The full-length UBAP1 protein is involved in endosomal dynamics in neurons, while loss of UBAP1 function may perturb endosomal fusion and sorting of ubiquitinated cargos. These effects could be more prominent in neurons, thereby giving rise to the phenotype of a neurodegenerative disease such as HSP.
Similar content being viewed by others
Introduction
Hereditary spastic paraplegias (HSPs) are a clinically and genetically heterogeneous group of rare neurodegenerative disorders characterized by progressive weakness and spasticity of the lower limbs [1]. They can be inherited in an autosomal dominant (AD), autosomal recessive (AR), X-linked pattern, or mitochondrial manner with an age of onset that varies from early childhood to 70 years of age [1,2,3]. Their estimated prevalence ranges from 1 to 10/100,000 depending on the geographic localization [4, 5]. To date, causative genes or gene loci for HSPs named SPG1 to SPG79 have been reported [6]. However; the genetic etiology remained unknown in ~40% of the HSP patients or families [7].
Ubiquitin-associated protein 1 (UBAP1), a subunit of mammalian endosomal sorting complex required for transport I (ESCRT-I), is coassembled in a stable 1:1:1:1 complex with TSG101, VPS28, and VPS37A [8,9,10], and is encoded by the UBAP1 gene [MIM: 609787]. The human and mouse UBAP1 proteins exhibit over 90% sequence identity. So far as we know, UBAP1 is part of a locus, where loss of heterozygosity frequently occurs in nasopharyngeal cancer [11]. Besides, some variants of UBAP1 were identified as risk factors in familial frontotemporal lobar dementia [12]. The importance of UBAP1 needs to be further underlined.
In this paper, we report six patients from four families with heterozygous truncating mutations in UBAP1 as the genetic cause of a new type of AD-HSP (SPG80). We created a mouse homolog of the mutation, and further characterized the UBAP1 wild type (WT) and UBAP1-mutant by examining their functional properties in mouse hippocampus neurons.
Material and methods
Patient recruitment and clinical evaluation
Six patients from four families were recruited and evaluated clinically by the Japan Spastic Paraplegia Research Consortium (JASPAC) [13], the University of Yamanashi, and the University of Tokyo. All patients showed spastic paraplegia as a pure form of juvenile-onset HSP (Table 1). The pedigree charts are shown in Fig. 1a. All patients and relatives gave their signed informed consent for this study. The Ethics Committee of the University of Yamanashi approved the project (Approval number: 734).
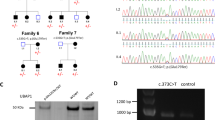
Pedigrees and sequence analysis of the juvenile-onset AD-HSP families. a Pedigrees of the juvenile-onset AD-HSP families we report. Squares indicate males; circles, females; slashes, deceased individuals; shaded (black) symbols, individuals with HSP; unshaded symbols, individuals without HSP; m/w, the heterozygous mutation in the UBAP1 gene from individuals with HSP; w/w, no mutation in the UBAP1 gene from individuals without HSP. b (a, b) Sequence analysis revealed the c.425_426delAG mutation in the UBAP1 gene in III-7 from Family #1, and II-6, III-2, and IV-1 from Family #2. The red arrow delineates the c.425 nucleotide. The green underline indicates conserved AG sequences in normal family members. c, d Sequence analysis revealed the c.312delC mutation in the UBAP1 gene in II-2 from Family #3 and the c.535G>T mutation in the UBAP1 gene in III-2 from Family #4. The red arrows delineate the c.312 nucleotide and c.535 nucleotide, respectively
Genetic analyses
We carried out whole-exome sequencing of genomic DNA from patients and both parents of patient III-7 in family #1. Genomic DNA was extracted from peripheral blood. Exome capture was performed with a SureSelect V5 + UTR (Agilent Technologies, CA, USA). Paired-end sequencing was carried out on a HiSeq2500 (Illumina, San Diego, CA, USA) using a HiSeq SBS Kit V4 (Illumina, San Diego, CA, USA), which generated 100-bp reads. We aligned the exome data with a Burrows–Wheeler Aligner and extracted single-nucleotide variations using SAMtools. We examined variants of known HSP genes through whole-exome analysis. The reference databases utilized included hg19 (GRCh37) (http://genome.ucsc.edu), ExAC (http://exac.broadinstitute.org), HGMD (https://portal.biobase-international.com), dbSNP, and the 1000 Genomes Project. Through screening variants of 87 genes known to be responsible for HSP or associated diseases (L1CAM, PLP2, ATL1, SPAST, CYP7B1, NIPA1, SPG7, KIAA0196, ALDH18A1, KIF5A, SPG11, RTN2, HSP60, ZFYVE26, BSCL2, ERLIN2, SPG20, ACP33, SLC16A2, RIPK5, B4GALNT1, DDHD1, KIF1A, REEP1, ZFYVE27, FA2H, PNPLA6, SLC33A1, C9orf12, GJC2, NT5C2, GBA2, AP4B1, AP5Z1, TECPR2, AP4M1, AP4E1, AP4S1, VPS37A, DDHD2, C12orf65, CYP2U1, TFG, KIF1C, USP8, WDR48, ARL6IP1, ERLIN1, AMPD2, ENTPD1, ARSI, PGAP1, KLC2, RAB3GAP2, MARS, ZFR, REEP2, CPT1C, IBA57, MAG, CAPN1, FARS2, ATP13A2, UCHL1, LYST, SACS, ABCD1, TUBB4A, ALS2, KCNA2, ABHD12, ANO10, CLCN1, CSF1R, GALC, GFAP, KCND3, MECP2, OPA1, PLA2G6, POLA3A, PSEN1, RNASEH1, SLC25A15, SYNE1, TYROBP, and WDR45), we narrowed down candidate variants among novel variants of all genes. We carried out Sanger sequencing of the six candidate genes. We referred to gnomAD (https://gnomad.broadinstitute.org), ToMMo (https://www.megabank.tohoku.ac.jp), and the Tokyo University in-house control database for normal control reference data.
Cloning of mouse UBAP1 cDNAs
Total RNA from the whole brains of male mice was extracted using QIAzol lysis reagent (Qiagen, Hilden Germany), and subsequently reverse-transcribed using a random primer (nonamer) and PrimeScrit II Reverse transcriptase (Takara Bio, Tokyo, Japan) according to the manufacturer’s instructions. Full-length mouse UBAP1 was PCR-amplified and subcloned into pBluescript II-KS (Stratagene, La Jolla, CA). Purified PCR products were analysed by DNA sequencing.
Molecular cloning of mouse UBAP1-mutant cDNAs
Mouse UBAP1-WT is full-length (amino acids 1–502), and mouse UBAP1-mutant (c.425_426delAG) chosen as our disease model is truncated (amino acids 1–157) according to gene model GenBank: NM_023305. We used pBluescript-UBAP1 as the template for PCR to prepare a construct of mouse UBAP1-mutant. The full-length fragment of mouse UBAP1-mutant (c.425_426delAG) was cloned from pBluescript-UBAP1-WT by PCR with the following oligonucleotides: pBS-mUBAP1-Forward: 5′-CGGGCTGCAGGAATTCATGGCTTCTAAGAAGTTGGG-3′; pBS-mUBAP1-Reverse: 5′-CCCCCTCGAGGTCGACTCAGCTGGCTCCAGCCCGGG-3′; UBAP1-425-426del-Forward: 5′-ACAAAGCAAAGTTCTCAGCCCACCC-3′; UBAP1-425-426del-Reverse: 5′-GAACTTTGCTTTGTGGCACTACTGC-3′. An EcoRI restriction site was included in the pBS-mUBAP1-Forward primer and a SalI restriction site in the pBS-mUBAP1-Reverse primer. Then the full-length fragment of mouse UBAP1-mutant was introduced using an In-Fusion cloning kit (Takara), and then subcloned into pCAII-EGFP for transfection.
Plasmid constructs
Expression vectors for UBAP1 and ubiquitin (Ub) were constructed in the pCAII-EGFP and pCAII-HA vectors, respectively, using standard molecular biological methods.
Primary neuron cultures and immunofluorescence image acquisition
Primary cultures of hippocampal neurons were prepared from 18-day-old pregnant Wistar rats as described previously [14]. On 11–13 days in vitro (DIV), neurons were transfected with the indicated expression vectors using Lipofectamine 2000 (Invitrogen). Immunostaining was performed at 13–15 DIV. The transfected neurons were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) (pH 7.4) for 30 min at room temperature, permeabilized with 0.25% Triton X-100 in PBS for 15 min, blocked with 0.25% Triton X-100 and 1% BSA in PBS for 30 min, and then incubated with primary antibodies diluted in the blocking solution for 2 h, followed by secondary antibodies for 1 h. Fluorescence images were acquired by confocal laser microscopy (Fluoview FV1200, Olympus, Tokyo, Japan) using a ×60 oil immersion objective lens. The fluorescence intensity was manually traced. Acquisition of microscopy images and morphometric quantification were performed by investigators blind as to the experimental conditions.
Antibodies
We used the following primary antibodies: anti-MAP2 (1:400, GeneTex); anti-GFP (1:500, Life Technologies); anti-Rab5 (1:200, Thermo Fisher); anti-Rab7 (1:100, Thermo Fisher); and anti-HA (1:400, Roche Applied Science). To detect the immunofluorescence signals, Alexa Fluor 488- or 568-labeled secondary antibodies (1:1000, Molecular Probes, Eugene, OR) were used. For Western blotting, horseradish peroxidase-linked secondary antibodies (1:2000-1:5000, GE Healthcare, Little Chalfont, UK) were used.
Western blotting
COS7 cells in 6 cm dishes were transfected with the indicated expression vectors using polyethylenimine. Then the cells were collected and lysed with boiling SDS-PAGE buffer, electrophoresed, and transferred to nitrocellulose membranes. After a blocking reaction (5% nonfat dry milk in Tris-buffered saline, pH 7.4, 0.05% Tween-20), primary antibodies (anti-GFP, 1:500) were incubated overnight at 4 °C in the blocking buffer. After incubation with a secondary antibody, an enhanced chemiluminescence procedure was performed.
Statistics
The UBAP1 distribution pattern was morphologically classified by investigators blind as to the transfected construct into “punctate”, “homogeneous distribution”, and “others”. For each observation, these rates were calculated. The observation data for three experiments were used collectively for statistical analysis. A two-way ANOVA and a post hoc Sidak’s multiple comparison test were used for classification of the intracellular distributions of UBAP1-WT and UBAP1-mutant. Statistical significance was assumed when p < 0.01.
Results
Gene analysis
There were 14 common variants (rs5490007670, rs10594016, rs2289189, rs775200011, rs13438232, rs10079250, rs398607, rs1126642, rs7624750, rs17849654, rs2306914, rs214950, rs2295191, and rs2295192), 11 out of the 87 genes are listed above, but no suspect variants in all four families.
In family #1, we identified five novel de novo variants in a heterozygous state in five genes (UBAP1, MUC12, C12orf55, GOLGA6L6, and ANKRD36C), and three novel variants in a compound heterozygous state in one gene (CNTNAP3B) in patient III-7 on exome sequencing. On Sanger sequencing, we reconfirmed only one de novo small deletion variant in the UBAP1 gene (Fig. 1b). Six other missense variants of the four genes (MUC12, C12orf55, ANKRD36C, and CNTNAP3B) were not reconfirmed and thus we judged them to be errors. It was difficult to confirm one variant of the GOLGA6L6 gene by Sanger sequencing due to the existence of huge analogous sequences. Therefore, we found only one likely candidate frameshift mutation (c.425_426del, p.K143Sfs*15) of the UBAP1 gene (GenBank: NM_016525.4).
After analysis of family #1, we screened the whole-exome sequencing data for 2376 index cases including 330 cases with HSP from the JASPAC database and another 2046 cases (1567 index cases with various neurological disorders from the Tokyo University in-house database, 315 cases with multiple system atrophy from the JAMSAC database, and 164 normal controls) to confirm that the UBAP1 gene is associated with HSP. By this means, we found additional one nonsense and two frameshift mutations in UBAP1 in the JASPAC database and Tokyo University in-house database, respectively. In addition, we found seven missense variants and one in-frame deletion variant in these databases (c.43A>G, c.107T>G, c.347C>T, c.670G>C, c.782G>A, c.989C>T, c.1328T>C, and c.1185_1187delTGT).
We detected the same frameshift mutation (c.425_426del, p.K143Sfs*15) of the UBAP1 gene in IV-1 of family #2, another frameshift mutation (c.312delC, p.S105Pfs*46) in II-2 of family #3, and a nonsense mutation (c.535G>T, p.E179*) in III-2 of family #4 from the exome sequencing data (Fig. 1b). We reconfirmed all three variants of the UBAP1 gene on Sanger sequencing. All four mutations are frameshift or nonsense ones, and all patients have common clinical pictures of the pure type of juvenile-onset HSP. In addition, we carried out Sanger sequencing of genomic DNA from another two patients and one unaffected relative in family #2 (II-6, III-2, and III-3) and confirmed that only the patients have the same mutation (Fig. 1b). Thus, these results would provide evidence for cosegregation. We could not obtain cooperation from the members of families #3 and #4.
Meanwhile, we judged all missense variants and an in-frame deletion to be noncausative variants for the following reasons. One variation (c.43A>G) was found in one HSP patient, one cerebellar ataxia patient and a healthy control. In addition, this variant was registered in ToMMo. Other variants were found in three HSP patients (c.670G>C, c.782G>A and c.1185_1187delTGT), three MSA patients (c.107T>G, c.989C>T and c.1328T>C), and one myopathy patient (c.347C>T). Among the three HSP patients, two show a complicated type and are SACS mutation carriers. In another patient the onset age was 49. These clinical features differ from those of the four families mentioned above with the pure type of juvenile-onset HSP.
As a result, we judged UBAP1 to be a novel causative gene of AD-HSP (SPG80) at least in a state of null mutation heterozygosity. The pathogenicity of other variants remains uncertain.
Protein function analysis
The UBAP1-mutant is a C-terminal deletion mutant
Mammalian UBAP1 protein interacts with Ub via its C-terminal SOUBA domain (residues 381–502), and utilizes its N-terminal UMA domain (residues 17–63) to bind the central stalk region of ESCRT-I. Besides, it contains a central region (residues 159–308) that binds ESCRT accessory protein HD-PTP (His domain protein tyrosine phosphatase; PTPN23) (Fig. 2a). HD-PTP regulates several Ub-dependent endosomal trafficking processes by binding to ESCRT subunits including UBAP1 [15,16,17].
The UBAP1-mutant is a C-terminal deletion mutant. a A schematic representation of the full-length UBAP1 protein and predicted effect on the protein structure of mutations in the four pedigrees on the basis of gene model GenBank: NM_016525.4. a Domain organization of the full-length UBAP1 protein. b–d The predicted effect in silico of mutations p.K143Sfs*15, p.S105Pfs*46 and p.E179* on the UBAP1 protein structure. b Western blot analysis indicates that the UBAP1-mutant is a C-terminal deletion mutant. pEGFP, pEGFP-UBAP1-WT or pEGFP-UBAP1-mutant was transfected into COS7 cells. Anti-GFP antibodies were used as primary antibodies. pEGFP was used as a loading control. Numbers indicate molecular mass in kDa. A specific band near 83.1 kDa was observed for GFP-UBAP1-WT extracts, and a specific band near 45.3 kDa for GFP-UBAP1-mutant extracts
All the UBAP1 mutations carried by the four pedigrees we report generated prematurely truncated proteins with complete loss of the SOUBA domain in the C-terminal region of UBAP1, the N-terminal UMA domains all being intact (Fig. 2a). As a result, the predicted consequences of these mutations will be a complete loss of Ub-binding function during endosomal sorting activities, and completely preserved efficiency of binding to the central stalk region of ESCRT-I. As Pedigrees 1 and 2 shared the same mutation (p.K143Sfs*15), which resulted in a premature stop codon at position 157 in both human and mouse UBAP1, we choose this mutation as the disease model and created a mouse c.425_426delAG homolog for a protein function study. We generated a plasmid encoding the truncated UBAP1 protein with a GFP (green fluorescent protein) tag fused to the N-terminal region. For Western blot analysis, the GFP-tagged UBAP1-WT and UBAP1-mutant plasmids were transfected into COS7 cells. GFP was also transfected as a loading control. Western blot analysis gave three specific bands with an anti-GFP antibody. The 83.1 kDa molecular weight band corresponds to GFP-UBAP1-WT, while the 45.3 kDa molecular weight one corresponds to GFP-UBAP1-mutant (Fig. 2b). This finding demonstrated truncated cytoplasmic protein expression of UBAP1-mutant. This C-terminal deletion UBAP1-mutant of our disease model has lost its Ub-binding domain, which is crucial for endosomal sorting of ubiquitinated cargos.
UBAP1 overexpression in primary cultured neurons
To investigate the functions of UBAP1-WT and UBAP1-mutant in neurons, we used an in vitro cell culture model that recapitulates key in vivo features of UBAP1. To determine if there was a systematic change in cell morphology in vitro, we transfected mouse hippocampal neurons in a dispersed cell culture with GFP-UBAP1-WT and GFP-UBAP1-mutant. The accuracy of identifying processes of hippocampal neurons was confirmed by double immunofluorescence with anti-MAP2 (a dendritic protein, which was a useful marker for identifying neurons in this study [18]). The hippocampus is a source of relatively homogeneous neurons exhibiting a stereotypical manner of differentiation in vitro, representing an ideal system to observe the morphological effects of various experimental conditions for neuron study [19].
As shown in Fig. 3a, both UBAP1-WT and UBAP1-mutant transfected mouse hippocampal neurons displayed a uniform size and shape. However, the overexpression of UBAP1-WT protein gave a punctate fluorescence pattern in hippocampal neurons. In contrast, the overexpression of UBAP1-mutant proteins gave a diffuse fluorescence pattern. This experiment was performed three times with reproducible similar results. A total of 214 neurons either transfected with UBAP1-WT or UBAP1-mutant were chosen blindly and randomly for statistical classification. These neurons were divided into three morphological patterns (Fig. 3b). The first pattern was named “punctate”, in which discrete speckles or brightly and distinctly clustered larger granules were seen within the cytoplasm. The nucleolar region was negative. This punctate pattern accounted for 70.60 ± 11.06% GFP-UBAP1-WT transfected neurons and 3.93 ± 1.98% GFP-UBAP1-mutant transfected neurons (p < 0.01). The second pattern was named “homogeneous distribution”, in which there was diffuse fluorescence of the entire neurons as well as the nucleus region with uniform intensity. This homogeneous distribution pattern accounted for 80.62 ± 7.42% UBAP1-mutant transfected neurons and 23.46 ± 10.58% UBAP1-WT transfected neurons (p < 0.01). The third pattern was named “others” due to an irregular morphology which was difficult to classify as either the first or second pattern. This pattern accounted for a very small proportion of neurons, which thus could be ignored. Thus, the morphology on the overexpression of UBAP1-mutant is quite different from that of UBAP1-WT in a primary culture of mouse hippocampal neurons. The overexpression of UBAP1-WT generates discrete puncta in the neuron cytoplasm, which is absent from UBAP1-mutant neurons. The overexpression of UBAP1-mutant gave homogeneous diffusion in the neuron cytoplasm and throughout the nucleus.
UBAP1 overexpression in primary cultured neurons. a The overexpression of GFP-UBAP1-WT and GFP-UBAP1-mutant gives a large punctate pattern and a diffuse pattern, respectively. Mouse hippocampal neurons were transfected with pEGFP-UBAP1-WT or pEGFP-UBAP1-mutant for 48 h and permeabilized before fixation. The cells were stained with a primary antibody against MAP2. DAPI fluoresces blue; GFP fluoresces green; MAP2 is dyed magenta. Scale bar, 20 μm. b Quantification was performed. The analysis of 214 neurons with each MAP2 marker gave the percentages shown in the graph. Error bars denote SEM
UBAP1 is colocalized with endosomes
The ESCRTs facilitate endosomal sorting of ubiquitinated cargos. ESCRT-0, -I, and -II capture ubiquitinated membrane proteins for sorting into intraluminal vesicles within endosomes to form structures known as multivesicular bodies [20]. During this process, UBAP1 as the subunit of ESCRT-I is recruited to endosome membranes [21, 22].
Rab5 small GTPase has been shown to be required for early endosome fusion and is used as an early endosome marker [23]. Rab7 small GTPase mediates late endosomal trafficking and fusion with autophagosomes, and thus is used as a late endosome marker [24]. To investigate the relationship between endosomes and our observation of UBAP1-positive puncta, GFP-UBAP1 transfected neurons were stained for the two above-mentioned markers, and then imaged by confocal microscopy. We found that GFP-UBAP1-WT-positive structures were colocalized in early endosomes, as labeled with the standard early endosome marker Rab5, and in late endosomes, based on Rab7 staining. In contrast, the diffuse fluorescence pattern of GFP-UBAP1-mutant overexpression was not colocalized with either marker of endosomes (Fig. 4). These studies revealed that the full-length UBAP1 protein is localized on expanded endosomes in the neuron cytoplasm. On the other hand, the UBAP1-mutant cannot be recruited to endosome membranes.
UBAP1 is colocalized with endosomes. The overexpressed UBAP1-WT puncta are costained with endosomal Rab5 and Rab7. Mouse hippocampal neurons were transfected with pEGFP-UBAP1-WT or pEGFP-UBAP1-mutant for 48 h and permeabilized before fixation. The cells were stained with a primary antibody against Rab5 or Rab7 and secondary antibodies as described under Materials and Methods. Control images of the GFP-transfected cells are shown in the left lane. DAPI fluoresces blue; GFP fluoresces green; Rab5/Rab7 is dyed magenta. Cyan in the merged images indicates colocalization. Scale bar, 20 μm
Overexpression of UBAP1-WT, but not mutant UBAP1, enhances ubiquitin association with endosomes in neurons
After endocytosis, some membrane proteins are recycled from early endosomes to the plasma membrane, whereas others are transported to late endosomes and lysosomes for degradation [25]. Conjugation with the small polypeptide Ub is a signal for endosome-lysosomal sorting [26, 27]. To investigate the Ub-binding functions of UBAP1-WT and UBAP1-mutant in mammalian neurons, mouse hippocampal neurons were cotransfected with HA-Ub and either UBAP1-WT or UBAP1-mutant for confocal immunofluorescence microscopy. We firstly cotransfected hippocampal neurons with HA-Ub and GFP, and then studied the localization of the exogenous Ub in the neuron cytoplasm. Neurons transfected with GFP, UBAP1-WT, or UBAP1-mutant alone were observed as control groups. As shown in Fig. 5, the exogenous Ub was distributed homogeneously around the nucleus in the neuron cytoplasm. Although the overexpression of UBAP1-mutant also exhibited a homogeneous distribution in the neuron cytoplasm, it is noteworthy that it could diffuse across the nuclear boundary. Therefore, UBAP1-mutant is unlikely to be colocalized with exogenous Ub, which is absent from the nucleus region in hippocampal neurons. On the other hand, exogenous Ub showed a punctate fluorescent pattern in hippocampal neurons cotransfected with UBAP1-WT. Significantly, in these cotransfected neurons we observed strong colocalization of UBAP1-WT and exogenous Ub. This experiment was performed three times with reproducible similar results. As noted earlier in the text, overexpression of UBAP1-WT is localized on expanded endosomes; we further demonstrated that UBAP1-WT interacts directly with Ub on endosomes in vitro, while UBAP1-mutant loses its ability to bind Ub as expected in silico.
The overexpressed UBAP1-WT is colocalized with ubiquitin, while UBAP1-mutant is not. Mouse hippocampal neurons transfected with pEGFP, pEGFP-UBAP1-WT, or pEGFP-UBAP1-mutant were cotransfected with HA-Ub for 48 h and permeabilized before fixation. Then the neurons were stained for confocal immunofluorescence microscopy with anti-HA (Ms) antibodies as described under “Materials and methods” section. Control images of the HA-Ub nontransfected cells are shown at the left in each lane. DAPI fluoresces blue; GFP fluoresces green; HA-Ub is dyed magenta. Cyan in the merged images indicates colocalization. Scale bar, 20 μm
Discussion
We report that heterozygous loss-of-function mutations in UBAP1 lead to HSP (SPG80). The patients’ common clinical features are juvenile-onset, slowly progressive, and a pure form of HSP with AD or de novo inheritance. According to our statistics based on the JASPAC data, HSP due to a UBAP1 mutation occurs at a frequency of 0.56% in all HSP patients and 1.5% in AD-HSP patients in Japan. At the same time, we found some heterozygous missense variants of UBAP1 in some neurodegenerative disease patients having family histories of HSP. However, they exhibit a variety of clinical features, such as spastic paraplegia, multiple system atrophy, or spinocerebellar ataxia. Therefore, evaluating the pathogenic effect of missense variants of UBAP1 will require further studies.
The functions of the proteins involved in HSPs are converging on a group of common pathogenic themes at the cellular level, such as axon pathfinding, myelination, endoplasmic reticulum network morphology, lipid metabolism, motor-based transport, mitochondrial function, and endosomal dynamics [6, 28]. To date, more HSP proteins have been implicated in endosomal dynamics than any other themes, including SPG4/spastin, SPG6/NIPA1, SPG8/strumpellin, SPG11/spatacsin, SPG15/spastizin, SPG20/spartin, SPG21/maspardin, SPG47/AP-4, SPG48/AP-5, SPG49/TECPR2, SPG53/VPS37A, SPG59/USP8, SPG60/WDR48, and SPG78/ATP13A2. Among them, SPG4 protein spastin and SPG20 protein spartin have been implicated in ESCRT-III protein. Spastin harbors a microtubule interacting and trafficking (MIT) domain that binds the ESCRT-III subunits CHMP1B and IST1, thereby coupling the severing of microtubules with membrane scission. These ESCRT interactions are crucial for spastin’s role in severing of microtubules to complete the abscission phase of cytokinesis [29,30,31]. It is worth mentioning that SPG4 is the most common AD HSP. Spartin harbors an MIT domain as well and interacts selectively with ESCRT-III subunit IST1 to participate in cytokinesis [32]. Although spastin and spartin have also been implicated in other pathogenic themes, one possible pathogenic mechanism for HSP is roles in the ESCRT-III interactions with spastin and spartin in the delivery and downregulation of cell surface receptors to regulate signaling in axons [28].
Moreover, another member of the ESCRT system, VPS37A, has been identified as the disease-causing gene for SPG53 [33]. The authors showed a homozygous missense mutation (pK382N) in VPS37A to be the cause of a novel form of complex AR-HSP. Although functional study of K382 in the VPS37A mod(r) domain was not performed, the authors speculated the regulation of protein expression through its binding to Ub. This binding signals protein degradation by lysosomes [34].
Therefore, dysfunction of spastin and spartin, which interact with ESCRT-III, as well as dysfunction of VPS37A and UBAP1, which are subunits of ESCRT-I, can point to a unified pathophysiology of ESCRT-dependent endosomal sorting activities.
As described in the introduction, UBAP1 is coassembled with TSG101, VPS28, and VPS37A to form ESCRT-I via binding to the central stalk region of ESCRT-I. This mammalian ESCRT-I central stalk happens to be composed of the VPS37A-TSG101 combination [9]. Hence, being the subunits of ESCRT-I, TSG101, VPS37A, and UBAP1 may exhibit some similarities in the genotype to phenotype relationship. Interestingly, the mutation of VPS37A causes complex AR-HSP, while the UBAP1 mutation we report is the genetic cause of pure AD-HSP. Besides, oligodendroglial deletion of TSG101 in mice causes spongiform encephalopathy, a fatal neurodegenerative disease [35].
In this study, we showed that the C-terminal deletion UBAP1-mutant of our disease model has lost its Ub-binding domain and therefore has lost its ability to bind Ub in vitro. The overexpression of UBAP1-WT interacts directly with Ub on enlarged endosomes in mouse hippocampal neurons, while the UBAP1-mutant cannot be recruited to endosome membranes. Therefore, we conclude that UBAP1-mutant loses its function of endosomal sorting of ubiquitinated cargos in neurons.
It is intriguing that the overexpression of the full-length UBAP1 protein induced enlarged Rab5-positive endosomes, suggesting that the UBAP1 protein functions in the early endosomal compartments in neurons. Conversely, the UBAP1-mutant cannot be recruited to endosomes. Rab5 is required for the fusion of early endosomes [23], and overexpression of the rab5 protein increases the size of early endosomes [36]. From the results of our experiment, we assume that UBAP1 overexpression in neurons stimulates Rab5-dependent endosomal fusion, resulting in enlarged endosomes. Coincidentally, a type of infantile onset HSP has been reported resulting from an AR mutation of the ALS2 gene encoding the alsin protein, a guanine nucleotide exchange factor for small GTPases Rab5 and Rac1 [27]. Rab5-dependent endosomal fusion is impaired in neurons from Als2 null mice, whereas alsin overexpression in neurons promotes Rab5-dependent endosomal fusion, giving rise to enlarged endosomes [37,38,39,40]. These phenotypic and functional similarities between ALS2 and UBAP1 further demonstrate that endosomal dynamics, including endosome trafficking and fusion, play crucial roles in the pathogeneses involving both genes in HSP.
Notably, the drastic enlargement of endosomes in cultured neuronal cells induced by overexpression of the full-length UBAP1 protein was not observed in COS7 cells, which were derived from monkey kidney tissue. Both overexpression of UBAP1-WT and UBAP1-mutant caused homogeneous diffusion in COS7 cells (Fig. S1). This suggests that neuronal cells might be more amenable to activation of UBAP1-associated endosomal dynamics, and thus the devastating effects due to the loss of the UBAP1functional domain could be more prominent in neurons. Such a neuron-specific effect of loss of UBAP1 function perturbs endosomal fusion and sorting activities, thereby giving rise to the phenotype of a neurodegenerative disease such as HSP.
During the preparing of submitting this article, truncating mutations in UBAP1 were reported in HSP families of diverse geographic origin [41]. Similarly, the majority of patients described in that report showed spastic paraplegia as a pure form of juvenile-onset HSP. In addition, they found features of cerebellar involvement in one family, and we report one patient with finger hyperextension.
In conclusion, we report mutations in the UBAP1 gene as the genetic cause of a new type of AD-HSP (SPG80). All the patients with novel heterozygous frameshift or nonsense mutations of the UBAP1 gene exhibited identical clinical features as a pure type of juvenile-onset AD-HSP or sporadic spastic paraplegia with a de novo mutation. Functional studies revealed UBAP1 is involved in endosomal dynamics in vitro. Loss of UBAP1 function may perturb endosomal fusion and sorting of ubiquitinated cargos in neurons, which may serve as the pathogenesis in HSP. Knockout mice for our disease model have been created. Further methods are being developed to enable study in vivo, and models will be introduced for new drug screening.
Accession code
The nucleotide sequence data reported are available in the GenBank. database under the accession numbers: MN137231, MN137232, MN137233, MN137234, MN137235, and MN137236.
References
Harding AE. Classification of the hereditary ataxias and paraplegias. Lancet. 1983;1:1151–5.
Tesson C, Koht J, Stevanin G. Delving into the complexity of hereditary spastic paraplegias: how unexpected phenotypes and inheritance modes are revolutionizing their nosology. Hum Genet. 2015;134:511–38.
Finsterer J, Löscher W, Quasthoff S, Wanschitz J, Auer-Grumbach M, Stevanin G. Hereditary spastic paraplegias with autosomal dominant, recessive, X-linked, or maternal trait of inheritance. J Neurol Sci. 2012;318:1–18.
Ruano L, Melo C, Silva MC, Coutinho P. The global epidemiology of hereditary ataxia and spastic paraplegia: a systematic review of prevalence studies. Neuroepidemiology. 2014;42:174–83.
Klebe S, Stevanin G. Clinical and genetic heterogeneity in hereditary spastic paraplegias: from SPG1 to SPG72 and still counting. Rev Neurol. 2015;171:505–30.
Blackstone C. Hereditary spastic paraplegia. Handb Clin Neurol. 2018;148:633–52.
Schüle R, Wiethoff S, Martus P, Karle KN, Otto S, Klebe S, et al. Hereditary spastic paraplegia: clinicogenetic lessons from 608 patients. Ann Neurol. 2016;79:646–58.
Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21:77–91.
Wunderley L, Brownhill K, Stefani F, Tabernero L, Woodman P. The molecular basis for selective assembly of the UBAP1-containing endosome-specific ESCRT-I complex. J Cell Sci. 2014;127:663–72.
Agromayor M, Soler N, Caballe A, Kueck T, Freund SM, Allen MD, et al. The UBAP1 subunit of ESCRT-I interacts with ubiquitin via a SOUBA domain. Structure. 2012;20:414–28.
Qian J, Yang J, Zhang X, Zhang B, Wang J, Zhou M, et al. Isolation and characterization of a novel cDNA, UBAP1, derived from the tumor suppressor locus in human chromosome 9p21-22. J Cancer Res Clin Oncol. 2001;127:613–8.
Rollinson S, Rizzu P, Sikkink S, Baker M, Halliwell N, Snowden J, et al. Ubiquitin associated protein 1 is a risk factor for frontotemporal lobar degeneration. Neurobiol Aging. 2009;30:656–65.
Koh K, Ishiura H, Tsuji S, Takiyama Y. JASPAC: Japan Spastic Paraplegia Research Consortium. Brain Sci. 2018;8:153.
Ohtsuka T, Takao-Rikitsu E, Inoue E, Inoue M, Takeuchi M, Matsubara K, et al. CAST: a novel protein of the cytomatrix at the active zone of synapses that forms a ternary complex with RIM1 and munc13-1. J Cell Biol. 2002;158:577–90.
Stefani F, Zhang L, Taylor S, Donovan J, Rollinson S, Doyotte A, et al. UBAP1 is a component of an endosome-specific ESCRT-I complex that is essential for MVB sorting. Curr Biol. 2011;21:1245–50.
Gahloth D, Levy C, Heaven G, Stefani F, Wunderley L, Mould P, et al. Structural basis for selective interaction between the ESCRT regulator HD-PTP and UBAP1. Structure. 2016;24:2115–26.
Ali N, Zhang L, Taylor S, Mironov A, Urbé S, Woodman P. Recruitment of UBPY and ESCRT exchange drive HD-PTP-dependent sorting of EGFR to the MVB. Curr Biol. 2013;23:453–61.
Caceres A, Banker G, Steward O, Binder L, Payne M. MAP2 is localized to the dendrites of hippocampal neurons which develop in culture. Brain Res. 1984;315:314–8.
Dotti CG, Sullivan CA, Banker GA. The establishment of polarity by hippocampal neurons in culture. J Neurosci. 1988;8:1454–86.
Shields SB, Piper RC. How ubiquitin functions with ESCRTs. Traffic. 2011;12:1306–17.
Gruenberg J, Stenmark H. The biogenesis of multivesicular endosomes. Nat Rev Mol Cell Biol. 2004;5:317–23.
Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–52.
Gorvel JP, Chavrier P, Zerial M, Gruenberg J. Rab 5 controls early endosome fusion in vitro. Cell. 1991;64:915–25.
Numrich J, Ungermann C. Endocytic Rabs in membrane trafficking and signaling. Biol Chem. 2014;395:327–33.
Gruenberg J. The endocytic pathway: a mosaic of domains. Nat Rev Mol Cell Biol. 2001;10:721–30.
Hicke L. Protein regulation by monoubiquitin. Nat Rev Mol Cell Biol. 2001;3:195–201.
Dupré S, Volland C, Haguenauer-Tsapis R. Membrane transport: ubiquitylation in endosomal sorting. Curr Biol. 2001;11:R932–R934.
Blackstone C. Cellular pathways of hereditary spastic paraplegia. Annu Rev Neurosci. 2012;35:25–47.
Yang D, Rismanchi N, Renvoisé B, Lippincott-Schwartz J, Blackstone C, Hurley JH. Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. Nat Struct Mol Biol. 2008;15:1278–86.
Connell JW, Lindon C, Luzio JP, Reid E. Spastin couples microtubule severing to membrane traffic in completion of cytokinesis and secretion. Traffic. 2009;10:42–56.
Guizetti J, Schermelleh L, Mäntler J, Maar S, Poser I, Leonhardt H, et al. Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science. 2011;331:1616–20.
Renvoise´ B, Parker RL, Yang D, Bakowska JC, Hurley JH, Blackstone C. SPG20 protein spartin is recruited to midbodies by ESCRT-III protein Ist1 and participates in cytokinesis. Mol Biol Cell. 2010;21:3293–303.
Zivony-Elboum Y, Westbroek W, Kfir N, Savitzki D, Shoval Y, Bloom A, et al. A founder mutation in Vps37A causes autosomal recessive complex hereditary spastic paraparesis. J Med Genet. 2012;49:462–72.
Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–6.
Walker WP, Oehler A, Edinger AL, Wagner KU, Gunn TM. Oligodendroglial deletion of ESCRT-I component TSG101 causes spongiform encephalopathy. Biol Cell. 2016;108:324–37.
Bucci C, Parton RG, Mather IH, Stunnenberg H, Simons K, Hoflack B, et al. The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell. 1992;70:715–28.
Devon RS, Orban PC, Gerrow K, Barbieri MA, Schwab C, Cao LP, et al. Als2-deficient mice exhibit disturbances in endosome trafficking associated with motor behavioral abnormalities. Proc Natl Acad Sci USA. 2006;103:9595–600.
Deng H-X, Zhai H, Fu R, Shi Y, Gorrie GH, Yang Y, et al. Distal axonopathy in an alsin-deficient mouse model. Hum Mol Genet. 2007;16:2911–2020.
Hadano S, Kunita R, Otomo A, Suzuki-Utsunomiya K, Ikeda JE. Molecular and cellular function of ALS2/alsin: implication of membrane dynamics in neuronal development and degeneration. Neurochem Int. 2007;51:74–84.
Otomo A, Hadano S, Okada T, Mizumura H, Kunita R, Nishijima H, et al. ALS2, a novel guanine nucleotide exchange factor for the small GTPase Rab5, is implicated in endosomal dynamics. Hum Mol Genet. 2003;12:1671–87.
Mohammad A, Adriana R, Elena B, Hamid N, Hassan D, Ina G, et al. Truncating mutations in UBAP1 cause hereditary spastic paraplegia. Am J Hum Genet. 2019;104:767–73.
Acknowledgements
We thank all the participants and cooperating doctors in the JASPAC. This work was supported by Grants-in-Aid from the Research Committee for Ataxic Disease (YT), the Ministry of Health, Labor and Welfare, Japan, JSPS KAKENHI Grant Number JP18K07495 (YT) from the Ministry of Education, Culture, Sports, Science, and Technology, Japan, and grants for AMED under Grant Number JP18kk0205001h003 (YT).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Nan, H., Ichinose, Y., Tanaka, M. et al. UBAP1 mutations cause juvenile-onset hereditary spastic paraplegias (SPG80) and impair UBAP1 targeting to endosomes. J Hum Genet 64, 1055–1065 (2019). https://doi.org/10.1038/s10038-019-0670-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-019-0670-9
This article is cited by
-
Ubap1 knock-in mice reproduced the phenotype of SPG80
Journal of Human Genetics (2022)
-
Challenges and Controversies in the Genetic Diagnosis of Hereditary Spastic Paraplegia
Current Neurology and Neuroscience Reports (2021)
-
Identification of UBAP1 mutations in juvenile hereditary spastic paraplegia in the 100,000 Genomes Project
European Journal of Human Genetics (2020)
-
Autosomal dominant hereditary spastic paraplegia caused by mutation of UBAP1
neurogenetics (2020)