Abstract
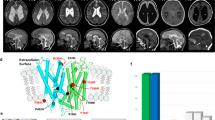
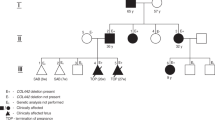
Heterozygous variants in CLTC, which encode the clathrin heavy chain protein, cause neurodevelopmental delay of varying severity, and often accompanied by dysmorphic features, seizures, hypotonia, and ataxia. To date, 28 affected individuals with CLTC variants have been reported, although their phenotypes have not been fully elucidated. Here, we report three novel de novo CLTC (NM_001288653.1) variants in three individuals with previously unreported clinical symptoms: c.3662_3664del:p.(Leu1221del) in individual 1, c.2878T>C:p.(Trp960Arg) in individual 2, and c.2430+1G>T:p.(Glu769_Lys810del) in individual 3. Consistent with previous reports, individuals with missense or small in-frame variants were more severely affected. Unreported symptoms included a brain defect (cystic lesions along the lateral ventricles of the brain in individuals 1 and 3), kidney findings (high-echogenic kidneys in individual 1 and agenesis of the left kidney and right vesicoureteral reflux in individual 3), respiratory abnormality (recurrent pneumonia in individual 1), and abnormal hematological findings (anemia in individual 1 and pancytopenia in individual 3). Of note, individual 1 even exhibited prenatal abnormality (fetal growth restriction, cystic brain lesions, high-echogenic kidneys, and a heart defect), suggesting that CLTC variants should be considered when abnormal prenatal findings in multiple organs are detected.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Briant K, Redlingshofer L, Brodsky FM. Clathrin’s life beyond 40: connecting biochemistry with physiology and disease. Curr Opin Cell Biol. 2020;65:141–9.
Brodsky FM. Diversity of clathrin function: new tricks for an old protein. Annu Rev Cell Dev Biol. 2012;28:309–36.
Kasprowicz J, Kuenen S, Miskiewicz K, Habets RL, Smitz L, Verstreken P. Inactivation of clathrin heavy chain inhibits synaptic recycling but allows bulk membrane uptake. J Cell Biol. 2008;182:1007–16.
Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alfoldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–43.
Nabais Sá MJ, Venselaar H, Wiel L, Trimouille A, Lasseaux E, Naudion S, et al. De novo CLTC variants are associated with a variable phenotype from mild to severe intellectual disability, microcephaly, hypoplasia of the corpus callosum, and epilepsy. Genet Med. 2020;22:797–802.
Hamdan FF, Myers CT, Cossette P, Lemay P, Spiegelman D, Laporte AD, et al. High rate of recurrent de novo mutations in developmental and epileptic encephalopathies. Am J Hum Genet. 2017;101:664–85.
DeMari J, Mroske C, Tang S, Nimeh J, Miller R, Lebel RR. CLTC as a clinically novel gene associated with multiple malformations and developmental delay. Am J Med Genet Part A. 2016;170A:958–66.
Manti F, Nardecchia F, Barresi S, Venditti M, Pizzi S, Hamdan FF, et al. Neurotransmitter trafficking defect in a patient with clathrin (CLTC) variation presenting with intellectual disability and early-onset parkinsonism. Parkinsonism Relat Disord. 2019;61:207–10.
Schmitz-Abe K, Li Q, Rosen SM, Nori N, Madden JA, Genetti CA, et al. Unique bioinformatic approach and comprehensive reanalysis improve diagnostic yield of clinical exomes. Eur J Hum Genet. 2019;27:1398–405.
Lecoquierre F, Duffourd Y, Vitobello A, Bruel AL, Urteaga B, Coubes C, et al. Variant recurrence in neurodevelopmental disorders: the use of publicly available genomic data identifies clinically relevant pathogenic missense variants. Genet Med. 2019;21:2504–11.
Bruel AL, Nambot S, Quere V, Vitobello A, Thevenon J, Assoum M, et al. Increased diagnostic and new genes identification outcome using research reanalysis of singleton exome sequencing. Eur J Hum Genet. 2019;27:1519–31.
Lelieveld SH, Reijnders MR, Pfundt R, Yntema HG, Kamsteeg EJ, de Vries P, et al. Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat Neurosci. 2016;19:1194–6.
Helbig KL, Farwell Hagman KD, Shinde DN, Mroske C, Powis Z, Li S, et al. Diagnostic exome sequencing provides a molecular diagnosis for a significant proportion of patients with epilepsy. Genet Med. 2016;18:898–905.
Aoi H, Mizuguchi T, Suzuki T, Makino S, Yamamoto Y, Takeda J, et al. Whole exome sequencing of fetal structural anomalies detected by ultrasonography. J Hum Genet. 2021;66:499–507.
Hosseini-Alghaderi S, Baron M. Notch3 in development, health and disease. Biomolecules. 2020;10:485.
Xu Y, Streets AJ, Hounslow AM, Tran U, Jean-Alphonse F, Needham AJ, et al. The polycystin-1, lipoxygenase, and α-toxin domain regulates polycystin-1 trafficking. J Am Soc Nephrol. 2016;27:1159–73.
Wilbur JD, Hwang PK, Ybe JA, Lane M, Sellers BD, Jacobson MP, et al. Conformation switching of clathrin light chain regulates clathrin lattice assembly. Dev Cell. 2010;18:841–8.
Digby EL, Liauw J, Dionne J, Langlois S, Nikkel SM. Etiologies and outcomes of prenatally diagnosed hyperechogenic kidneys. Prenat Diagn. 2021;41:465–77.
Harambat J, van Stralen KJ, Kim JJ, Tizard EJ. Epidemiology of chronic kidney disease in children. Pediatr Nephrol. 2012;27:363–73.
Acknowledgements
We thank all the participants in this study. We are grateful to Ms. M. Satoh, Mr. T. Miyama, Ms. K. Takabe, Ms. N. Watanabe, and Ms. S. Sugimoto from the Department of Human Genetics, Yokohama City University Graduate School of Medicine for their technical assistance. This work was supported by the Agency for Medical Research and Development (AMED) under grant numbers JP21ek0109486, JP21ek0109549, JP21cm0106503, and JP21ek0109493 (NM); Japan Society for the Promotion of Science (JSPS) KAKENHI under grant numbers, JP20K07907 (SM), JP20K08164 (TM), JP20K17936 (AF), JP20K16932 (KH), JP19K17865 (YU), and JP20K17428 (NT); and the Takeda Science Foundation (TM and NM). We thank Rachel James, Ph.D., from Edanz Group (https://en-author-services.edanz.com/ac) for editing a draft of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Itai, T., Miyatake, S., Tsuchida, N. et al. Novel CLTC variants cause new brain and kidney phenotypes. J Hum Genet 67, 1–7 (2022). https://doi.org/10.1038/s10038-021-00957-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-021-00957-3
This article is cited by
-
A novel de novo CLTC variant altering RNA splicing causes fetal developmental abnormalities
BMC Medical Genomics (2023)