Abstract
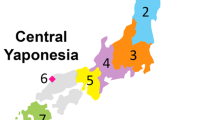
We analyzed genome-wide single-nucleotide polymorphism data of 11,069 Japanese individuals recruited from all 47 prefectures of Japan to clarify their genetic structure. The principal component analysis at the prefectural level enabled us to study the relationship between geographical location and genetic differentiation. The results revealed that the mainland Japanese were not genetically homogeneous, and the genetic structure could be explained mainly by the degree of Jomon ancestry and the geographical location. One of the interesting findings was that individuals in the Shikoku region (i.e., Tokushima Prefecture, Kagawa Prefecture, Ehime Prefecture, and Kochi Prefecture) were genetically close to Han Chinese. Therefore, the genetic components of immigrants from continental East Asia in the Yayoi period may have been well maintained in Shikoku. The present results will be useful for understanding the peopling of Japan, and also provide suggestions for recruiting subjects in genetic association studies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Hanihara K. Dual structure model for the population history of the Japanese. Jpn Rev. 1991;2:1–33.
Hanihara K. Geographic variation of modern Japanese crania and its relationship to the origin of Japanese. Homo. 1985;36:1–10.
Kanzawa-Kiriyama H, Kryukov K, Jinam AT, Hosomichi K, Saso A, Suwa G, et al. A partial nuclear genome of the Jomons who lived 3000 years ago in Fukushima, Japan. J Hum Genet. 2017;62:213–21.
Kanzawa-Kiriyama H, Jinam AT, Kawai Y, Sato T, Hosomichi K, Tajima A, et al. Late Jomon male and female genome sequences from the Funadomari site in Hokkaido, Japan. Anthropol Sci. 2019;1–26. https://doi.org/10.1537/ase.190415.
Hammer MF, Horai S. Y chromosomal DNA variation and the peopling of Japan. Am J Hum Genet. 1995;56:951–62.
Horai S, Murayama K, Hayasaka K, Matsubayashi S, Hattori Y, Fucharoen G, et al. mtDNA polymorphism in East Asian populations, with special reference to the peopling of Japan. Am J Hum Genet. 1996;59:579–90.
Hatta Y, Ohashi J, Imanishi T, Kamiyama H. HLA genes and haplotypes in Ryukyuans suggest recent gene flow to the Okinawa Islands. Hum Biol. 1999;71:353–65.
Yamaguchi-Kabata Y, Nakazono K, Takahashi A, Saito S, Hosono N, Kubo M, et al. Japanese population structure, based on SNP genotypes from 7003 individuals compared to other ethnic groups: effects on population-based association studies. Am J Hum Genet. 2008;83:445–56.
Japanese Archipelago Human Population Genetics Consortium. The history of human populations in the Japanese Archipelago inferred from genome-wide SNP data with a special reference to the Ainu and the Ryukyuan populations. J Hum Genet. 2012;57:787–95.
Hellwege JN, Keaton J, Giri A, Gao X, Velez Edwards DR, Edwards TL. Population stratification in genetic association studies. Curr Protoc Hum Genet. 2017;95:1.22.1–23.
Berg JJ, Harpak A, Sinnott-Armstrong N, Joergensen AM, Mostafavi H, Field Y, et al. Reduced signal for polygenic adaptation of height in UK biobank. Elife. 2019;8:1–47.
Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4.
1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526:68–74.
Wickham H. ggplot2: elegant graphics for data analysis. New York, NY: Springer; 2016.
Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–64.
Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25.
Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, et al. Ancient admixture in human history. Genetics. 2012;192:1065–93.
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–9.
Jinam TA, Kanzawa-Kiriyama H, Inoue I, Tokunaga K, Omoto K, Saito N. Unique characteristics of the Ainu population in Northern Japan. J Hum Genet. 2015;60:565–71.
Koganebuchi K, Kimura R. Biomedical and genetic characteristics of the Ryukyuans: demographic history, diseases and physical and physiological traits. Ann Hum Biol. 2019;46:354–66.
Acknowledgements
We are grateful to the individuals who participated in the study. We would like to express our deepest gratitude to Masahiro Inoue, Shota Arichi, and Akito Tabira who obtained the genotype data and provided the technical environment for analyzing them. We thank two anonymous reviewers for helpful suggestions. This study was partly supported by Grant-in-Aid for Scientific Research (B) (18H02514) and Grant-in-Aid for Scientific Research on Innovative Areas (19H05341) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and supported by AMED under Grant Numbers JP19fk0310115 and JP20km0405211.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Watanabe, Y., Isshiki, M. & Ohashi, J. Prefecture-level population structure of the Japanese based on SNP genotypes of 11,069 individuals. J Hum Genet 66, 431–437 (2021). https://doi.org/10.1038/s10038-020-00847-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-020-00847-0
This article is cited by
-
Genetic diversity among the present Japanese population: evidence from genotyping of human cell lines established in Japan
Human Cell (2024)
-
Demographic history of Ryukyu islanders at the southern part of the Japanese Archipelago inferred from whole-genome resequencing data
Journal of Human Genetics (2023)
-
Influence of the nutritional status on facial morphology in young Japanese women
Scientific Reports (2022)
-
Geographic variation in the polygenic score of height in Japan
Human Genetics (2021)