Abstract
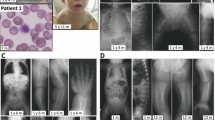
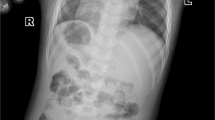
NGLY1 deficiency is the first and only autosomal recessive congenital disorder of N-linked deglycosylation (NGLY1-CDDG). To date, no patients with NGLY1 deficiency has been reported from mainland China or East Asia in English literature. Here, we present six patients with a diagnosis of NGLY1-CDDG on the basis of clinical phenotype, genetic testing, and functional studies. We retrospectively analyzed clinical phenotypes and NGLY1 genotypes of six cases from four families. Informed consent was obtained for diagnosis and treatment. In-silico tools and in vitro enzyme activity assays were used to determine pathogenicity of NGLY1 varaints. All patients had typical features of NGLY1-CDDG, including global developmental delay, microcephaly, hypotonia, hypertransaminasemia, alacrimia, and feeding difficulty. Dysmorphic features found in our patients include flat nasal bridge, loose and hollow cheeks, short stature, malnutrition, and ptosis. Pachylosis could be a novel cutaneous feature that may be explained by lack of sweat. We found three novel variants, including one missense (c.982C > G/p.Arg328Gly), one splice site (c.1003+3A > G), and one frame-shift (c.1637-1652delCATCTTTTGCTTATAT/p.Ser546PhefsTer) variant. All mutations were predicted to be disease causing with in-silico prediction tools, and affected at least one feature of gene splicing. Protein modeling showed missense variants may affect covalent bonding within the protein structure, or interrupt active/binding amino-acid residues. In vitro studies indicated that proteins carrying missense variants (p.Arg328Gly and p.Tyr342Cys) lost the enzyme activity. We expanded clinical phenotype and genetic mutation spectrum of NGLY1-CDDG by reporting six cases, three novel variants, and novel clinical features from mainland China.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Suzuki T, Huang C, Fujihira H. The cytoplasmic peptide: N-glycanase (NGLY1)-Structure, expression and cellular functions. Gene. 2016;577:1–7.
Need AC, Shashi V, Hitomi Y, Schoch K, Shianna KV, McDonald MT, et al. Clinical application of exome sequencing in undiagnosed genetic conditions. J Med Genet. 2012;49:353–61.
Enns GM, Shashi V, Bainbridge M, Gambello MJ, Zahir FR, Bast T, et al. Mutations in NGLY1 cause an inherited disorder of the endoplasmic reticulum-associated degradation pathway. Genet Med. 2014;16:751–8.
Caglayan AO, Comu S, Baranoski JF, Parman Y, Kaymakçalan H, Akgumus GT, et al. NGLY1 mutation causes neuromotor impairment, intellectual disability, and neuropathy. Eur J Med Genet. 2015;58:39–43.
He P, Grotzke JE, Ng BG, Gunel M, Jafar-Nejad H, Cresswell P, et al. A congenital disorder of deglycosylation: Biochemical characterization of N-glycanase 1 deficiency in patient fibroblasts. Glycobiology. 2015;25:836–44.
Heeley J, Shinawi M. Multi-systemic involvement in NGLY1-related disorder caused by two novel mutations. Am J Med Genet A. 2015;167A:816–20.
Lam C, Ferreira C, Krasnewich D, Toro C, Latham L, Zein WM, et al. Prospective phenotyping of NGLY1-CDDG, the first congenital disorder of deglycosylation. Genet Med. 2017;19:160–8.
Zhi Li, Liu F. [NGLY1 gene mutation causes congenital disorder of glycosylation type IV: a family report.]. J Clin Pediatr. 2018;35:904–7. http://www.wanfangdata.com.cn/details/detail.do?_type=perio&id=lcekzz201812005 (article in Chinese)
Wang L, Yu X, Li M, Sun G, Zou L, Li T, et al. Filamentation initiated by Cas2 and its association with the acquisition process in cells. Int J Oral Sci. 2019;11:29.
Sun G, Yu X, Bao C, Wang L, Li M, Gan J, et al. Identification and characterization of a novel prokaryotic peptide: N-glycosidase from Elizabethkingia meningoseptica. J Biol Chem. 2015;290:7452–62.
Hou L, Li T, Chen H, Shen D, Li M, Guo Y, et al. Identification and characterization of a novel glycoprotein core xylosidase from the bacterium Elizabethkingia meningoseptica. Biochem Biophys Res Commun. 2019;517:390–7.
Li T, Li M, Hou L, Guo Y, Wang L, Sun G, et al. Identification and characterization of a core fucosidase from the bacterium Elizabethkingia meningoseptica. J Biol Chem. 2018;293:1243–58.
Waisbren SE, He J, McCarter R. Assessing Psychological functioning in metabolic disorders: validation of the adaptive behavior assessment system, second edition (ABAS-II), and the behavior rating inventory of executive function (BRIEF) for identification of individuals at risk. JIMD Rep. 2015;21:35–43.
Ryu SW, Kim E. Apoptosis induced by human Fas-associated factor 1, hFAF1, requires its ubiquitin homologous domain, but not the Fas-binding domain. Biochem Biophys Res Commun. 2001;286:1027–32.
Tickotsky-Moskovitz N. New perspectives on the mutated NGLY1 enigma. Med Hypotheses. 2015;85:584–5.
Galeone A, Han SY, Huang C, Hosomi A, Suzuki T, Jafar-Nejad H. Tissue-specific regulation of BMP signaling by Drosophila N-glycanase 1. Elife. 2017 Aug 4;6. pii: e27612.
Maerz S, Funakoshi Y, Negishi Y, Suzuki T, Seiler S. The Neurospora peptide: N-glycanase ortholog PNG1 is essential for cell polarity despite its lack of enzymatic activity. J Biol Chem. 2010;285:2326–32.
Altrich-VanLith ML, Ostankovitch M, Polefrone JM, Mosse CA, Shabanowitz J, Hunt DF, et al. Processing of a class I-restricted epitope from tyrosinase requires peptide N-glycanase and the cooperative action of endoplasmic reticulum aminopeptidase 1 and cytosolic proteases. J Immunol. 2006;177:5440–50.
Kario E, Tirosh B, Ploegh HL, Navon A. N-linked glycosylation does not impair proteasomal degradation but affects class I major histocompatibility complex presentation. J Biol Chem. 2008;283:244–54.
van Keulen BJ, Rotteveel J, Finken MJJ. Unexplained death in patients with NGLY1 mutations may be explained by adrenal insufficiency. Physiol Rep. 2019;7:e13979.
Cahan EM, Frick SL. Orthopaedic phenotyping of NGLY1 deficiency using an international, family-led disease registry. Orphanet J Rare Dis. 2019;14:148.
Allen MD, Buchberger A, Bycroft M. The PUB domain functions as a p97 binding module in human peptide N-glycanase. J Biol Chem. 2006;281:25502–8.
Zhou X, Zhao G, Truglio JJ, Wang L, Li G, Lennarz WJ, et al. Structural and biochemical studies of the C-terminal domain of mouse peptide-N-glycanase identify it as a mannose-binding module. Proc Natl Acad Sci USA. 2006;103:17214–9.
Zhao G, Li G, Zhou X, Matsuo I, Ito Y, Suzuki T, et al. Structural and mutational studies on the importance of oligosaccharide binding for the activity of yeast PNGase. Glycobiology. 2009;19:118–25.
Hall PL, Lam C, Alexander JJ, Asif G, Berry GT, Ferreira C, et al. Urine oligosaccharide screening by MALDI-TOF for the identification of NGLY1 deficiency. Mol Genet Metab. 2018;124:82–6.
Haijes HA, de Sain-van der Velden MGM, Prinsen HCMT, Willems AP, van der Ham M, Gerrits J, et al. Aspartylglycosamine is a biomarker for NGLY1-CDDG, a congenital disorder of deglycosylation. Mol Genet Metab. 2019 Jul 9. pii: S1096-7192(19)30360-9.
Chang CA, Wei XC, Martin SR, Sinasac DS, Al-Hertani W. Transiently elevated plasma methionine, S-adenosylmethionine and S-adenosylhomocysteine: Unreported laboratory findings in a patient with NGLY1 deficiency, a congenital disorder of deglycosylation. JIMD Rep. 2019;49:21–9.
Xiong X, Liu D, Wang Y, Zeng T, Peng Y. Urinary 3-(3-hydroxyphenyl)-3-hydroxypropionic acid, 3-hydroxyphenylacetic acid, and 3-hydroxyhippuric acid are elevated in children with autism spectrum disorders. Biomed Res Int. 2016;2016:9485412.
Huang C, Harada Y, Hosomi A, Masahara-Negishi Y, Seino J, Fujihira H, et al. Endo-β-N-acetylglucosaminidase forms N-GlcNAc protein aggregates during ER-associated degradation in Ngly1-defective cells. Proc Natl Acad Sci USA. 2015;112:1398–403.
Fujihira H, Masahara-Negishi Y, Tamura M, Huang C, Harada Y, Wakana S, et al. Lethality of mice bearing a knockout of the Ngly1-gene is partially rescued by the additional deletion of the Engase gene. PLoS Genet. 2017;13:e1006696.
Bi Y, Might M, Vankayalapati H, Kuberan B. Repurposing of proton pump inhibitors as first identified small molecule inhibitors of endo-β-N-acetylglucosaminidase (ENGase) for the treatment of NGLY1 deficiency, a rare genetic disease. Bioorg Med Chem Lett. 2017;27:2962–6.
Funding
This work was supported by National Natural Science Foundation of China [81873543, and 81570468 to J.S.W.] and National Science and Technology Major Project [2014ZX09101046-004 to L.C.].
Author information
Authors and Affiliations
Contributions
WJS and CL designed the study and approved the final submission; AK collected clinical and genetic data, performed protein modeling, conducted literature search, and summarized relevant information. Both WJS and AK clinically managed patients. ZL, WL, and CL performed functional studies. AK, ZL, and WL co-wrote the manuscript, and contributed equally for this study.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Abuduxikuer, K., Zou, L., Wang, L. et al. Novel NGLY1 gene variants in Chinese children with global developmental delay, microcephaly, hypotonia, hypertransaminasemia, alacrimia, and feeding difficulty. J Hum Genet 65, 387–396 (2020). https://doi.org/10.1038/s10038-019-0719-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-019-0719-9
This article is cited by
-
NGLY1 deficiency: estimated incidence, clinical features, and genotypic spectrum from the NGLY1 Registry
Orphanet Journal of Rare Diseases (2022)
-
Reversibility of motor dysfunction in the rat model of NGLY1 deficiency
Molecular Brain (2021)