Abstract
The human leukocyte antigen (HLA) genes are the most polymorphic genes in the human genome and have great power in forensic applications, especially in relationship testing and personal identification. However, the extreme polymorphism of HLA has made unambiguous genotyping of these genes very challenging and resulted in the limited application in relationship testing. Fortunately, massively parallel sequencing (MPS) technology offers the promise of unambiguous and high-throughput HLA typing. In this study, 11 HLA genes were typed in one extended family residing in North China and encompassing six generations. Phase-resolved genotypes for HLA genes were generated and HLA haplotype structure was defined. The paternity/kinship index, or in other words, likelihood ratio (LR) was calculated. A total of 88 alleles were identified, of which eight alleles were newly discovered. The inheritance of HLA alleles followed Mendelian law. With the discovery of new HLA alleles and three recombination events, a total of eleven new HLA haplotypes were identified in this population. LR distribution showed that, when HLA alleles were applied, the Log10LR for a single locus could reach very high and the median average Log10LRs of HLA genes were much higher than that of short tandem repeat loci. The result showed that high-throughput HLA genotyping could be achieved rapidly by MPS, and the contribution of HLA genes on system performance could be high, which may be applied as a supplement in forensic genetics studies. This study was also valuable in demonstrating the genetic mechanisms governing the generation of polymorphisms of the HLA genes.
Similar content being viewed by others
Introduction
The human leukocyte antigen (HLA) genes encoded by major histocompatibility complex (MHC) are known to be the most polymorphic genes in the human genome. They are located on the short arm of chromosome 6 in the band 6p 21.3 and classified into the HLA Class I, class II, and class III genes [1]. In addition to high polymorphism, the HLA genes are also characterized by a haplotypic inheritance feature. In general, they are not only pivotal in immune response, but also important for the diagnosis and prognosis of cancer, autoimmunity, and infectious disease [2, 3]. Besides, highly polymorphic HLA genes can also be used as important genetic markers in forensic applications, which was widely adopted in forensic paternity testing from the 1980s to the 1990s [4,5,6].
Owing to extensive allelic diversity, genotyping of HLA genes is quite challenging. Although techniques like sequence-specific oligonucleotide probes (SSOP), sequence-specific primer (SSP), and sequence-based typing have replaced serological methods and become routine HLA-typing methods [7], they are limited in throughput and precision, and only the antigen recognition site encoding exons are sequenced for most of the samples. Accordingly, they are incapable of resolving the two main types of ambiguity in HLA typing [7, 8]. The first exists when a heterozygous sequence can be explained by more than one possible pair of alleles within the analyzed region. The second results from polymorphisms outside the region being typed. Thus, typing of HLA genes was laborious and time-consuming and usually could not meet the demand of unambiguous HLA typing in forensic applications. As a result, with the rapid development and maturity of capillary electrophoresis (CE)-based short tandem repeat (STR) typing technology, HLA has gradually fallen into obscurity in the forensic application in the 21st century.
Although STRs are widely used in forensic DNA analysis at present, some limitations still exist. The main one is the relatively high mutation rates of STRs [9]. In kinship analysis, the combined paternity index (CPI) or combined kinship index (CKI) may not reach the required thresholds owing to the mutations of the studied markers, and additional STRs should be utilized. But additional STR markers may still be insufficient. The workload and cost are not low as well. Whether there is a supplement to currently used STR markers in the case of paternity disputes needs to be explored. Although the system performance of HLA alleles was thought as high in relationship testing for many years, no study made a comparison between HLA and STRs in a more comprehensive way.
In recent years, the introduction of massively parallel sequencing (MPS) has revolutionized HLA-genotyping methods [10]. MPS can realize large-scale parallel sequencing of clonal amplified single deoxyribonucleic acid (DNA) molecules within a few days, which makes it suitable for high-throughput HLA typing and resolving the ambiguity problem [11,12,13,14]. In addition, some groups have successfully leveraged the throughput power of MPS to achieve unambiguous HLA typing with relatively low cost through sample multiplexing [13, 15,16,17]. Application of MPS-based HLA typing in the forensic field may facilitate reintroduction of HLA markers in forensic applications. The potential power of HLA genes on relationship testing in the era of new sequencing technology needs to be explored. The TruSight HLA assay was proved to enable high-throughput HLA typing with high accuracy, low ambiguity rate, and low cost [18]. It can generate phase-resolved sequencing results for the most intensely studied HLA genes including HLA-A, -C, -B, -DQA1, -DQB1, -DPA1, -DPB1, -DRB1, -DRB3, -DRB4, and -DRB5. Here, 24 Chinese Han individuals, belonging to a family residing in North China and spanning six generations, were studied using the Illumina MiSeq platform.
Materials and methods
DNA sample
One donor family from the northern Chinese Han population, containing 24 individuals, was recruited (see Supplementary Figure 1 for the pedigree structure). Whole blood samples were collected from these 24 samples. All samples were collected with written consent from the anonymous donors, who gave their permission for DNA analyses and scientific publication. This study was approved by the Ethics Committee of Sun Yat-sen University.
DNA extraction
Genomic DNA was extracted using the QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA), and the total DNA was quantified using the Qubit 3.0 Fluorometer with the Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific) following the manufacturer’s instructions. After quantification, the sample DNA was diluted to 10 ng/μL each.
STR typing
All the samples were genotyped using the Goldeneye DNA ID System 25 A amplification system (Peoplespot SciTech Incorporation, Beijing, China), which includes 20 expanded Combined DNA Index System (CODIS) core loci plus three loci (Penta E, Penta D, and D6S1043), as well as a Y indel and Amelogenin loci for sex identification. DNA amplification was performed according to the manufacturer’s instructions. The polymerase chain reaction (PCR) products were detected with CE on an ABI3500 Genetic Analyzer (Applied Biosystems, USA). The results were analyzed with the GeneMapper ID-X Analysis Software (Applied Biosystems).
HLA gene amplification, library preparations, and sequencing
HLA genes were amplified using a TruSight HLA v2 Sequencing Panel (Illumina, Inc., San Diego, CA). Long-range PCR for the 11 loci was performed in eight separate PCR reactions (HLA-A, -C, -B, -DQA1, -DQB1, -DPA1, -DPB1, and -DRB1/3/4/5). Fifty nanograms of genomic DNA were used for amplification of each HLA locus. PCR was performed according to the manufacturer’s instructions. PCR amplicons were visualized using 0.8% agarose gel electrophoresis before preparing MPS libraries. Sequencing libraries were prepared according to the manufacturer’s instructions. Twenty-four barcoded samples (192 HLA libraries) were pooled and quantified using a Qubit 3.0 fluorometer. The pooled HLA library was loaded onto the cartridge and sequenced using MiSeq Reagent Kit v2 Micro (300 cycles) on Illumina MiSeq platform. Demultiplexing and generation of FASTQ files were performed on the MiSeq system.
Data analysis
Raw sequencing data in FASTQ format were analyzed using TruSight HLA Assign 2.0 Software (version 2.1.0.943) supplied by Illumina. The software aligned the individual reads to a consensus reference sequence and phased all heterozygous positions using base positions under individual reads and paired reads. These phased alignments generate a consensus sequence for each locus, which was compared with known HLA allele sequences from the IMGT/HLA database (version 3.26), and the HLA genotype was assigned. If no allele with a 100% match to the known reference sequences was assigned, the mismatches with known reference sequences throughout the entire amplified region of the HLA genes were further analyzed. Mismatches that represented potential novel HLA alleles were noted and validated later.
The allele coverage ratios (ACR) were calculated for heterozygous samples. ACR was calculated by dividing the coverage of the lower coverage allele by that of the higher coverage allele.
The paternity/kinship index was calculated for each pair of individuals by comparing the “related” vs. “unrelated” hypotheses [19]. The paternity/kinship index is a likelihood ratio (LR) that is generated by comparing two probabilities. In brief, for a specific relationship, the model compared the likelihood value (L) based on the genotypes of involved individuals for the following two hypotheses: H0, the tested person was the specific member in this relationship; H1, the tested person was an unrelated person. Then the LR value was calculated as the ratio of L (G|H0) and L(G|H1), where G refers to genotypes. The formulas for calculation of LRs were provided in Supplementary Table 1. The allele frequencies of the HLA alleles were collected from the Allele Frequency Net Database (http://www.allelefrequencies.net/default.asp) and Zhou’s study [20]. For the 23 autosomal STRs in Goldeneye™ DNA ID System 25 A, the allele frequencies and locus-specific mutation rates were collected from the STRBase (http://www.cstl.nist.gov/strbase/). For HLA genes, the frequency of higher field allele was applied for alleles without frequency data (e.g., if no frequency data were available for B*33:03:01, the frequency of B*33:03 was applied). LR for those cases with suspect mutations was calculated according to Technical specifications for paternity testing issued by the Ministry of Justice of the People’s Republic of China. The combined LR (CLR) was calculated by multiplying LRs based on the product rule. In this study, an LR of more than 10,000 (i.e., Log10LR of 4) was considered as proof of parent–offspring relationship. For the comparison analysis of the theoretical impact on system performance of STR loci and HLA genes, three different pairwise relations including parent–offspring, full siblings, second-degree relatives (i.e., grandparent–grandchild, uncle/aunt–nephew/niece) were analyzed.
Sanger sequencing
For putative new alleles, the single-nucleotide polymorphisms (SNPs) detected in the MPS method were further validated by Sanger sequencing. All samples with putative new alleles were amplified using ExTaq DNA polymerase (Takara Inc., Dalian, China). In individuals who were heterozygous for a locus, the primers were designed at the flanking heterozygous region to specifically amplify the SNP in the new alleles. After amplification, the amplicons were sequenced on an automated ABI3730xl Genetic Analyzer using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA). All procedures were performed following the manufacturer’s instructions.
Results
STR typing results
With the DNA ID System 25 A amplification system, two mutations were detected within the 15 trio families (Supplementary Table 2). The first one was a one-step mutation at FGA, where genotypes were heterozygote “23.2, 25” and “22, 25” for the father and the mother, but was “22, 26” for the child. It was a paternal mutation from 25 to 26. The second mutation was a maternal one-step mutation at D3S1358, with the father and the mother exhibit genotypes of “16, 17” and “14, 15” while the child exhibit “16, 17”. Both mutations have been confirmed by massively parallel sequencing in a later study (unpublished data).
HLA amplification, library preparation, and sequencing
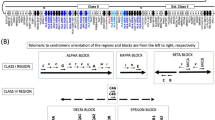
The HLA loci were amplified by long-range PCR and the analyzed regions are shown in Fig. 1 (a). Eleven HLA loci were amplified using eight long-range PCR reactions. Although the entire genes were studied for HLA-A, -C, -DPA1, -DQA1, and -DQB1, the partial sequences were analyzed for the remaining genes (-B, -DPB1, and -DRB1/3/4/5), with the studied regions covering most of the gene sequences. On the agarose gel, the bands of fragments HLA-DPA1, -DPB1, and -DRB1/3/4/5 were weak (Supplementary Figure 2). Subsequent library constructions were performed separately, and each HLA library was quantified after library preparation. As determined by the Qubit, the DNA library concentration ranged from 7.6 ng/μL to 31.2 ng/μL. The final pooled library was 15.9 ng/μL, and 0.95 μL of the pooled library was further sequenced.
a Outline of targeted PCR regions in eleven HLA loci. Black boxes indicate exons, and amplified regions are indicated using arrows. b The raw amplicon read counts for eight amplicons (nine genes/gene sets) are shown in boxplots. Boxes represent the median and the first and third quartiles. Whiskers correspond to the interquartile range, and outliers are plotted
The 24 individuals were sequenced in a single run (2 × 150). A total of 1503.6 MB data were obtained using MiSeq sequencing with cluster density was 949 K/mm2 and the clusters passing the filter was 90.8%. The average library fragment size was 422 base pair (bp). The distribution of varying fragments for the whole library shows that most (80.2%) reads were within the range of 200 bp to 600 bp (Supplementary Figure 3). The overall read quality and depth of coverage were determined for each HLA locus. The average percentage of reads ≥ Q30 was 94.1% (95% CI, 94.0–94.3%) for all HLA loci (Supplementary Figure 4). The average depth of coverage was 234.6× (95% CI, 231.8 × –237.5×) for all HLA loci (Fig. 1b).
HLA-typing result
At the field 4 level, a total of 88 unique alleles were identified by the TruSight HLA panel in the 24 samples (Table 1). The HLA-typing result for each sample was also shown in Supplementary Table 3. Of the 88 alleles, 80 were identical to the allele sequences recorded in the IMGT/HLA database. The remaining eight alleles were found to be putative novel alleles with high similarity to the reported sequences, but they contained SNP variants (Table 2). For the eight putative novel alleles, all the variants were observed in the intronic region. But one of the novel variants located on the intron 1/exon 2 boundary (see the HLA-DRB4 of sample 19 in Table 2). Five novel alleles appeared in more than one sample and followed the heredity rule (Table 2 and Fig. 2). For example, a new allele named A*02:01:01:28, with one base difference in intron 6 compared to A*02:01:01:01, was found in sample 4. This new allele was also observed in her two children and one grandson.
HLA alleles and HLA haplotypes in this family. HLA allele was inferred by IMG/HLA database and shared between parents and child(ren) with a consistent pattern. Three recombination events were observed and are indicated by black stars below the corresponding haplotypes. HLA-DRB3/4/5 were not included in the haplotype analysis, owing to limited available data
To validate the putative novel alleles, Sanger sequencing was performed, and the results confirmed all identified mutation sites. All sequences of new HLA alleles identified here have been deposited in GenBank under accession numbers MG571441 to MG571448. Five of these have been assigned the official name by the World Health Organization (WHO) Nomenclature Committee. The remaining three new alleles could be assigned official names if full-length sequences are obtained in the future.
Owing to the length limitation of PCR amplification, only a partial sequence was analyzed for some loci and the region containing variants that differentiate similar allele pairs may not be typed. Thus, some HLA alleles identified here may have two or more perfect matches to the reference sequences, creating an ambiguous result referred to as ambiguities. Of the 88 unique HLA alleles, 11 ambiguities were found, with a 100% match to more than one known alleles. For a specific locus, all matched ambiguities belong to the same HLA G group (Supplementary Table 3), and these alleles have identical nucleotide sequences across the exons encoding the peptide binding domains. Ambiguities frequently occurred in HLA-DQA1. No ambiguities were detected in the HLA-A, -C, -DPA1, -DQB1, -DRB3, -DRB4, and -DRB5 locus.
Allelic imbalance of the heterozygous samples was analyzed by calculating the ACR. As seen from Supplementary Figure 5, except for loci HLA-DRB1, the ACRs at most HLA loci were over 0.6, indicating that the allele pairs were overall balanced. This is a demonstration of the good performance of this HLA amplification system.
HLA haplotype structure
The phase-defined HLA haplotype structures were inferred by the family structure based on MPS genotyping results (Fig. 2). HLA-DRB3/4/5 were not included in the haplotype analysis, owing to limited available data. The result showed that all HLA genes were inherited following Mendelian inheritance. Of the 30 meiosis from 15 trio families, three recombination events were observed. The two paternal crossover recombination events occurred between HLA-A and -C, -B, and -DRB1, respectively. Sample 8 inherited a maternal haplotype that was the result of a recombination event between the mother’s HLA-B and -DRB1, which is also a crossover recombination.
There is a total of 21 different HLA haplotypes in this family, with three of them were generated by recombination events. As some HLA alleles were newly identified in this population, the corresponding HLA haplotypes should also be new within this population. With the addition of three recombinant HLA haplotypes, 11 HLA haplotypes are probably new haplotypes in this population.
Distribution of the LR
First, the parental relationship for the 15 trio families (30 parents and child pair) was determined by calculating the CLR value based on 23 autosomal STRs. Of the 30 pairs of parent–offspring duos, the Log10CLR of two pairs were below the threshold 4. One of which is attributed to the mutation observed in locus D3S1358 for sample 11 and 15. This inconclusive result could be solved by adding only one HLA locus either HLA-B or -DRB1. For the second case, adding any of the analyzed HLA loci except HLA-A could increase the CLR ratio.
The theoretical impacts on system performance were analyzed by calculating the LR for the studied STR loci and HLA-A, -C, -B, -DRB1, and -DQB1. Each allele collected in our study for the above loci was assumed as a shared allele by the two involved individuals and LR was calculated. Although the calculation formulas vary for a different degree of relationship, the distributions of Log10LR were similar because all the formula could be related to reciprocal of the frequency of the involved allele (Fig. 3). Thus, the distribution of Log10LR for only one case was shown in the figure. The result indicated that for all the five HLA gene, the Log10LR could reach very high, even when the rare alleles (alleles that appeared only once in this population) were excluded from the analysis. The median average Log10LRs of HLA genes were much higher than that of the STR loci.
The distribution of Log10LR when assuming each allele collected in our study for HLA genes and STR loci as shared allele by the two involved individuals. The formulas for calculating LRs are shown. Although the calculation formulas varied for the different degree of relationships, the distribution of Log10LR was similar because all the formula could be related to reciprocal of the frequency of the involved allele. Only the distribution pattern for parent–offspring pairs with heterozygous genotypes sharing one allele (gray background) was shown. Gene names with gray backgrounds indicate the results after eliminating the rare alleles (alleles that appeared only once in this population)
Discussion
Typing of HLA genes has been a challenge because conventional methods (Sanger sequencing, SSP, and SSOP) usually generate ambiguities that are time-consuming and expensive to resolve [21]. Massively parallel sequencing could reduce the ambiguity problem significantly when used in conjunction with the long-range PCR method. In this study, we demonstrated that the TruSight HLA assay allows rapid, accurate, high-throughput HLA genotyping, which is largely consistent with previous results [13, 15,16,17]. Still, of the HLA alleles identified here, eleven were ambiguities with at least two perfect matches (100% match) to the reference alleles. All the matched reference alleles for a specific ambiguity belong to the same HLA G group, that is, have identical nucleotide sequences across the exons. Thus, the ambiguities were speculated to have been caused by typing of partial sequences rather than the entire sequences. Analysis of a partial sequence may miss the SNPs that differentiate similar alleles. Further sequencing of polymorphisms outside the analyzed region should be able to resolve the ambiguous results. Thus, ambiguous typing prompts the need for full-length sequencing to increase the HLA-typing accuracies. This will not only help eliminate ambiguities but also allow the detection of DNA sequence polymorphisms that form putative new alleles.
For amplification efficiency of this method, the fragments with long amplicon sizes, such as HLA-DPA1 and -DPB1, and those with mixed gene products, such as HLA-DRB1/3/4/5, were the most challenging HLA genes to amplify. However, the sequencing result showed that the coverage was sufficient for accurate HLA assignment. Only one allele in HLA-DRB1, that is DRB1*04:05:01, had coverage lower than 200 reads and forms the outliers in Fig. 1b, which implies the low amplification efficiency of this allele. In addition, allelic imbalances were observed in some HLA loci within this panel, especially in HLA-B, -DQB1, and -DRB1 (Supplementary Figure 5). The most likely reason for the allele coverage bias is amplification imbalance. Amplification efficiency for specific allele seems low. For HLA-B, allele imbalances were observed in the heterogeneous sample with B*40:06:01:01 or B*39:01:01 G, whereas for HLA-DRB1, all the samples with amplification imbalance contain DRB1*04:05:01. For HLA-DQB1, all the allelic imbalances were observed in the heterogeneous samples with one allele belongs to allele group 03 or 04 and the other allele belongs to allele group 05 (allele group 03 means HLA-DQB1*03:xx:xx:xx). Therefore, although an absolute balanced allele coverage was impossible, further optimization of primers and primer pool are needed.
The HLA genes are characterized by high polymorphism and haplotypic inheritance features. The complexity of the HLA system is not only reflected by the extreme polymorphism of HLA genes but also the high polymorphism of HLA haplotypes. Among many molecular genetic mechanisms, point mutations and meiotic recombinations (gene conversion and crossing over) are key contributors to the generation of new polymorphisms [22]. Mutation in a specific HLA gene will generate new alleles and new HLA haplotypes, which will be transferred to the offspring. At the same time, recombination could also play a role in the generation of novel alleles at various HLA loci and is further responsible for the diversity observed at the haplotype level [23, 24]. In this study, several new HLA alleles were identified, which formed corresponding new HLA haplotypes. With the addition of three recombinant HLA haplotypes, a total of 11 HLA haplotypes were newly discovered in this population. All the recombination events occurred at previously reported recombination hotspots, namely, between HLA-A and -C, -B, and -DRB1 [23, 25].
Generation of new HLA alleles and HLA haplotypes may function positively in the adaptive evolution of the immune system by presenting parasite-derived antigens, which may play a critical role in maintaining population stability or in adapting to a new environment [26, 27]. It also increases the polymorphism of the HLA system, which would give the greatest expected discrimination power in human identity testing or relationship testing. Furthermore, as the HLA system was reported to contain conserved polymorphic regions that are shuffled by recombination to form ethnic-specific haplotypes [28], some recombination may help to generate informative haplotypes to differentiate the populations.
In forensic applications, HLA could become a powerful supplement to currently used STR markers in the case of paternity disputes or in discerning complex kinship relationships. First, although STRs are the most widely used genetic markers in forensic DNA analysis for the past decades, many studies have revealed that the relatively high mutation rates of STRs are a disadvantage for both human identification and relationship testing [9]. When the CPI or CKI was lower than the required thresholds either owing to the mutations or low efficiency of the studied markers, additional STRs should be included, which will help to confirm the relationship in inconclusive cases. But, even though additional STR markers could be used, the result may not be sufficient. The workload and cost are high as well. Under this circumstance, a highly polymorphic and heterozygous locus will become a powerful supplementary to the STR multiplex. Therefore, highly polymorphic HLA genes might be of special use in DNA testing. As has been shown in Fig. 3, when HLA alleles were applied, the Log10LR could reach very high and the median average Log10LRs of HLA genes are much higher than STR loci. Thus, the corresponding contribution on system performance will also be high. One locus of the HLA marker might significantly increase the value of CPI when alleles with low frequency are involved. Second, as many diseases are correlated with HLA alleles, the typing of HLA alleles may offer additional information on externally visible characteristics, which could become clues for criminal investigation.
In this study, HLA genes were typed at a high-resolution level in a big family from North China spanning six generations. The result showed that rapid and high-resolution HLA genotyping for many individuals at multiple HLA loci could be achieved simultaneously. Eight novel alleles were identified, and five of them have been assigned the official names by the World Health Organization (WHO) Nomenclature Committee [29,30,31]. The inheritance of HLA alleles followed Mendelian law. It not only confirmed the reliability of the sequencing results but also validated the putative new HLA alleles. In addition, LR distribution analysis showed that, the system performance of HLA alleles was overall higher than that of STR loci in relationship testing. Therefore, highly polymorphic HLA genes could become a powerful supplement to currently used STR markers in the case of paternity disputes or in discerning complex kinship relationships.
References
Guillaudeux T, Janer M, Wong GK, Spies T, Geraghty DE. The complete genomic sequence of 424,015 bp at the centromeric end of the HLA class I region: gene content and polymorphism. Proc Natl Acad Sci USA. 1998;95:9494–9.
Trowsdale J, Knight JC. Major histocompatibility complex genomics and human disease. Annu Rev Genom Hum Genet. 2013;14:301–23.
Opelz G, Wujciak T, Döhler B, Scherer S, Mytilineos J. HLA compatibility and organ transplant survival. Collaborative Transplant Study. Rev Immunogenet. 1999;1:334–42.
Grubic Z, Stingl K, Martinez N, Palfi B, Brkljacic-Kerhin V, Kastelan A. STR and HLA analysis in paternity testing. Int Congr Ser. 2004;1261:535–7.
Terasaki PI, Gjertson D, Bernoco D, Perdue S, Mickey MR, Bond J. Twins with two different fathers identified by HLA. N Engl J Med. 1978;299:590–2.
Kereszturya L, Rajczya K, Lászikb A, Gyódia E, Pénzes M, Falus A, et al. Combination of DNA-based and conventional methods to detect human leukocyte antigen polymorphism and its use for paternity testing. Am J Forensic Med Pathol. 2002;23:57–62.
Erlich H. HLA DNA typing: past, present, and future. Tissue Antigens. 2012;80:1–11.
Adams SD, Barracchini KC, Chen D, Robbins F, Wang L, Larsen P, et al. Ambiguous allele combinations in HLA Class I and Class II sequence-based typing: when precise nucleotide sequencing leads to imprecise allele identification. J Transl Med. 2004;2:30.
Brinkmann B, Klintschar M, Neuhuber F, Hühne J, Rolf B. Mutation rate in human microsatellites: influence of the structure and length of the tandem repeat. Am J Hum Genet. 1998;62:1408–15.
Lind C, Ferriola D, Mackiewicz K, Heron S, Rogers M, Slavich L, et al. Next-generation sequencing: the solution for high-resolution, unambiguous human leukocyte antigen typing. Hum Immunol. 2010;71:1033–42.
Bentley G, Higuchi R, Hoglund B, Goodridge D, Sayer D, Trachtenberg EA, et al. High-resolution, high-throughput HLA genotyping by next-generation sequencing. Tissue Antigens. 2009;74:393–403.
Gabriel C, Danzer M, Hackl C, Kopal G, Hufnagl P, Hofer K, et al. Rapid high-throughput human leukocyte antigen typing by massively parallel pyrosequencing for high-resolution allele identification. Hum Immunol. 2009;70:960–4.
Ehrenberg PK, Geretz A, Baldwin KM, Apps R, Polonis VR, Robb ML, et al. High-throughput multiplex HLA genotyping by next-generation sequencing using multi-locus individual tagging. BMC Genomics. 2014;15:864.
Hosomichi K, Jinam TA, Mitsunaga S, Nakaoka H, Inoue I. Phase-defined complete sequencing of the HLA genes by next-generation sequencing. BMC Genomics. 2013;14:355.
Shiina T, Suzuki S, Ozaki Y, Taira H, Kikkawa E, Shigenari A, et al. Super high resolution for single molecule-sequence-based typing of classical HLA loci at the 8-digit level using next generation sequencers. Tissue Antigens. 2012;80:305–16.
Ozaki Y, Suzuki S, Kashiwase K, Shigenari A, Okudaira Y, Ito S, et al. Cost-efficient multiplex PCR for routine genotyping of up to nine classical HLA loci in a single analytical run of multiple samples by next generation sequencing. BMC Genomics. 2015;16:318.
Duke JL, Lind C, Mackiewicz K, Ferriola D, Papazoglou A, Gasiewski A, et al. Determining performance characteristics of an NGS-based HLA typing method for clinical applications. HLA . 2016;87:141–52.
Weimer ET, Montgomery M, Petraroia R, Crawford J, Schmitz JL. Performance Characteristics and Validation of Next-Generation Sequencing for Human Leucocyte Antigen Typing. J Mol Diagn. 2016;18:668–75.
Wenk RE, Traver M, Chiafari FA. Determination of sibship in any two persons. Transfusion. 1996;36:259–62.
Zhou XY, Zhu FM, Li JP, Mao W, Zhang DM, Liu ML, et al. High-resolution analyses of human leukocyte antigens allele and haplotype frequencies based on 169, 995 volunteers from the China bone marrow donor registry program. PLoS ONE. 2015;10:1–26.
Yin Y, Lan JH, Nguyen D, Valenzuela N, Takemura P, Bolon Y-T, et al. Application of high-throughput next-generation sequencing for HLA typing on buccal extracted DNA: results from over 10,000 donor recruitment samples. PLoS ONE. 2016;11:e0165810.
Agrawal S, Srivastava S, Tripathi P. Role of Human Leukocyte Antigens in Studying Population Diversity. In: Bhasin V, Bhasin MK (eds). Anthropology today: trends, scope and applications. Kamla-Raj Enterprises: Delhi, India, 2007, pp 355–65.
Carrington M. Recombination within the human MHC. Immunol Rev. 1999;167:245–56.
Lebedeva TV, Huang A, Ohashi M, Sibilia P, Alosco SM, Kempenich J, et al. The recombinant HLA-B*5518 allele supports the evidence of conserved haplotype association of rare alleles. Tissue Antigens. 2005;66:156–9.
Miretti MM, Walsh EC, Ke X, Delgado M, Griffiths M, Hunt S, et al. A high-resolution linkage-disequilibrium map of the human major histocompatibility complex and first generation of tag single-nucleotide polyamorphisms. Am J Hum Genet. 2005;76:634–46.
Lenz TL, Spirin V, Jordan DM, Sunyaev SR. Excess of deleterious mutations around HLA genes reveals evolutionary cost of balancing selection. Mol Biol Evol. 2016;33:2555–64.
Petersdorf EW. The major histocompatibility complex: a model for understanding graft-versus-host disease. Blood. 2013;122:1863–72.
Gaudieri S, Leelayuwat C, Tay GK, Townend DC, Dawkins RL. The major histocompatability complex (MHC) contains conserved polymorphic genomic sequences that are shuffled by recombination to form ethnic-specific haplotypes. J Mol Evol. 1997;45:17–23.
Wu R, Li R, Zhang Y, Li H, Sun H. The novel HLA-DQB1*03:01:01:12 allele, identified by next-generation sequencing in a Chinese family. HLA. 2018;91:221–2.
Wu R, Li H, Wang N, Peng D, Sun H. Next-generation sequencing-based typing of a new allele HLA-DQB1*03:01:01:20 in a Chinese family. HLA. 2018;91:223–4.
Wu R, Peng D, Li R, Li H, Sun H. HLA-A*02:01:01:28, a novel HLA allele identified by next-generation sequencing in a Chinese family. HLA. 2018;91:195–6.
Acknowledgements
We are grateful to all volunteers who provided the samples used in this study. This study was funded by the National Natural Science Foundation of China (81801878), the Natural Science Foundation of Guangdong Province (2018A030313413) and the Fundamental Research Funds for the Central Universities (17ykpy10).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Wu, R., Li, H., Peng, D. et al. Revisiting the potential power of human leukocyte antigen (HLA) genes on relationship testing by massively parallel sequencing-based HLA typing in an extended family. J Hum Genet 64, 29–38 (2019). https://doi.org/10.1038/s10038-018-0521-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-018-0521-0
This article is cited by
-
Fine human genetic map based on UK10K data set
Human Genetics (2022)