Abstract
Spinocerebellar ataxia 37 (SCA37) is caused by an (ATTTC)n insertion in a polymorphic ATTTT repeat in the non-coding region of DAB1. The non-pathogenic alleles have a configuration [(ATTTT)7–400], whereas pathogenic alleles have a complex structure of [(ATTTT)60–79(ATTTC)31–75(ATTTT)58–90]. Molecular diagnosis of SCA37 is laborious because about 7% of the pentanucleotide repeat alleles in DAB1 are larger than 30 units and, thus, fail to amplify with standard PCR conditions, resulting in apparently homoallelism or in complete lack of PCR amplification in several cases. The molecular test currently available requires long-range PCR and sequencing analysis for the detection and characterization of these large alleles. We developed a simple assay capable of rapidly detecting the presence or absence of large pentanucleotide repeat sizes. This assay is based on repeat-primed PCR followed by high-throughput capillary electrophoresis. Combining the standard PCR with RP-PCR allows completion of the diagnosis in more than 80% of individuals, minimizing the number of samples that require long-range PCR followed by Sanger sequencing analysis. This assay meets many of the requirements for pre-screening of large cohorts of affected individuals.
Similar content being viewed by others
Introduction
A large number of neurological and neuromuscular diseases are caused by unstable repetitive tracts in coding or non-coding gene regions [1,2,3]. These include the expansion of trinucleotide repeats in coding regions, the most common being CAG repeats that encode toxic polyglutamine proteins [4, 5]. Tri-, tetra-, penta-, and hexanucleotide repeats in non-coding gene regions, however, are also responsible for many of these disorders [6,7,8,9,10,11]. In two of the unstable repeat diseases, spinocerebellar ataxia type 31 (SCA31) and SCA37, there is a pentanucleotide repeat insertion in another polymorphic pentanucleotide repeat, which is a challenge for routine diagnostic testing [12, 13]. This is further complicated by the existence of very large non-pathogenic alleles comprised of pentanucleotide repeats [13, 14].
In SCA37, there is an (ATTTC)n insertion in the middle of a polymorphic ATTTT repeat in the 5′UTR region of DAB1 (DAB1, reelin adaptor protein) [13]. The non-pathogenic alleles have ATTTT repeats that can vary from 7 to 400 units and never contain ATTTC. Standard PCR amplification of the pentanucleotide repeat in DAB1 can easily detect alleles shorter than 30 units [13]; however, amplification of larger alleles is technically challenging and requires long-range PCR [13]. By standard PCR, finding only one band of potentially two normal alleles of the same size (homoallelism) is not conclusive because there is the possibility of preferential amplification of the shorter allele in the presence of a large allele that is not amplified. As the molecular diagnosis relies on the presence of the ATTTC insertion and since 7% of non-pathological alleles are larger than 30 ATTTTs [13], long-range PCR and sequencing analysis are frequently required.
To facilitate screening of large cohorts of patients with SCA, there is a need for simple and inexpensive tests with high sensitivity and specificity. Expanded alleles in several of the SCAs and in fragile-X syndrome can be detected using a fluorescent repeat-primed PCR assay [15, 16]. In SCA37, the (ATTTC)n insertion has been successfully detected by a similar repeat-primed PCR adapted to the specificity of the pentanucleotide repeat insertion [13]. DNA samples with detected ATTTC repeat insertions are, then, amplified by long-range PCR followed by sequencing analysis of the products obtained. This method is adequate for repeat insertions flanked by ATTTT stretches of ~100 units; however, for much larger ATTTT flanking repeats the ladder of peaks obtained would fall out of the electropherogram detection.
For screening purposes, this limitation of the ATTTC RP-PCR negatively affects the sensitivity of the test. To overcome this problem, we have developed an ATTTT RP-PCR that allows detection of pentanucleotide repeat alleles in DAB1 larger than 30 units. With this screening method both normal alleles larger than 30 ATTTTs and alleles with the ATTTC repeat insertion flanked by ATTTTs are detected. The final step is always sequencing analysis of the products of long-range PCR to identify alleles with the ATTTC repeat insertion.
Subjects and methods
DNA samples from affected individuals and controls
This study used the de-identified, previously collected, DNA samples of affected individuals with SCA37 as well as anonymized DNA samples from control individuals; they were described in a previous study [13].
Repeat-primed PCR for the ATTTT repeat
The pentanucleotide repeat sequence in DAB1 was amplified by ATTTT RP-PCR with primers 24F (6-FAM-5′-GAAGTGGTCCTCCCAAGTCA-3′), FLAG (5′-TACGCATCCCAGTTTGAGACG-3′), and RP-AATAA (5′-TACGCATCCCAGTTTGAGACGAATAAAATAAAATAAAATAAAATAA-3′). The forward primer (24F) was a locus-specific primer, while the RP-AATAA primer was an ATTTT-repeat specific. The RP-AATAA primer had a DNA tail sequence at the 5′ end (absent in the human genome) and the reverse primer FLAG had the 5′ tail sequence of the RP-AATAA primer. PCR was performed with 100 ng genomic DNA, 1× HotStarTaq Master Mix (Qiagen), 0.8 µM primer 24F and primer FLAG, and 0.4 µM primer RP-AATAA, in 12.5 µL. The initial RP-PCR step was at 95 °C for 15 min and was followed by 40 cycles (94 °C for 1 min, 54 °C for 1 min, and 72 °C for 2 min and 30 s) and a final extension of 10 min at 72 °C. RP-PCR products were detected by capillary electrophoresis of 3 µl of RP-PCR product mixed with 0.5 µl of GS500(-250)LIZ size standard in Hi-DiTM formamide (Applied Biosystems). Samples were denatured at 95 °C, for 5 min, before loading onto an ABI3730xl DNA Analyzer on a 36-cm capillary. Fragment sizes were analyzed with GeneMapper v.4.0 (Applied Biosystems).
Sequencing analysis
The long-range PCR [13] was performed in duplicate for each sample with positive detection by ATTTT RP-PCR; PCR products were separated by electrophoresis in a 1% agarose gel. For each sample, each DNA and corresponding duplicate band (of 0.9 kb or larger) were sliced from the gel and the DNA was extracted together using the QIAquick gel extraction kit (Qiagen) following the manufacturer instructions. The DNA was analyzed by Sanger sequencing. Sequencing analysis in the forward orientation used 2 µL of DNA extracted from the duplicate bands together, 4 µL of BigDye terminator v.1.1 (Applied Biosystems), 0.5 × BigDye sequencing buffer, 2.5 µM of primer 24 F (5′-GAAGTGGTCCTCCCAAGTCA-3′) in a final volume of 20 µl. Sequencing analysis in the reverse orientation used 2 µl of DNA, 8 µL of BigDye terminator v.1.1, 0.5 × BigDye sequencing buffer and 1.5 µM of primer 24R4 (5′-GAGACCAGCCTGGGCAAC-3′) in a final volume of 20 µl. Sequencing reactions were performed, for both forward and reverse orientations, with an initial denaturation of 5 min at 95 °C followed by 50 cycles of 30 s at 95 °C, 10 s at 56 °C, and 4 min at 60 °C and a final extension of 10 min at 60 °C.
Results
RP-PCR of pentanucleotide repeat alleles
Standard PCR analysis of the pentanucleotide repeat in DAB1 resulted in the amplification of two, one or no alleles in samples from affected or control individuals (Table 1); this method could not amplify successfully alleles larger than 30 repeats [13]. To detect pentanucleotide repeats larger than 30 units in DAB1 for the screening of SCA37 in large cohorts of individuals, we developed a simple ATTTT RP-PCR (Fig. 1). After capillary electrophoresis and electropherogram analysis, we observed that alleles larger than 30 repeats produced a ladder of multiple peaks 5-bp apart and extending beyond the 250 bp, while this ladder ended below 250 bp for alleles smaller than 30 repeats. Thus, we predefined a cut-off value of 250 bp below which the test was considered negative, indicating the absence of the SCA37 pentanucleotide repeat insertion. All the samples with long non-pathogenic ATTTT tracts of 33–400 (17 samples) or with the pathogenic repeat insertion (41 samples) showed an ATTTT RP-PCR ladder extending beyond 250 bp.
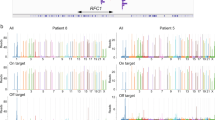
ATTTT RP-PCR to detect large pentanucleotide alleles in DAB1. a Schematic representation of the ATTTT RP-PCR primers that anneal with the repetitive ATTTT region, resulting in DNA amplification in normal and mutant alleles. b Electropherograms showing the fluorescent ATTTT RP-PCR analysis in control individuals from Table 1: C-75, C-88, C-91, C-95, and C-44; and in SCA37 affected individuals A-1 and A-9
Assay validation and specificity
One hundred and thirty seven samples previously analyzed by sequence analysis [13] were tested to validate the ATTTT RP-PCR assay. The samples showed full concordance with previous results from sequencing analysis (Table 1 and Fig. 2). All samples with a negative result by ATTTT RP-PCR had pentanucleotide repeat alleles shorter than 30, demonstrating the specificity of the assay (Fig. 1).
Sanger sequencing analysis of DAB1 pentanucleotide repeat alleles. a Short (ATTTT)8 allele and the repeat flanking region, previously amplified with standard PCR. b Electropherogram fragment of an (ATTTT)n allele with more than 30 repeat units, amplified using long-range PCR. c Electropherogram fragments of an allele with the repeat insertion from an affected individual with SCA37. Each fragment shows repeat motifs that constitute the allele and are positioned according to the structure [(ATTTT)n(ATTTC)n(ATTTT)n]. The arrows represent the continuation of the repeat stretch; the double red lines represent the (ATTTC)n insertion
Discussion
Diagnostic testing for the ATTTC repeat insertion in DAB1 that causes SCA37 requires amplification of a complex (ATTTT)n(ATTTC)n(ATTTT)n sequence of approximately 200 pentanucleotides and its sequencing analysis [13]. To further complicate this test, non-pathogenic alleles may be composed of >30 ATTTT repeats in 7% of the population and, thus, not be amplified with standard PCR conditions, showing complete lack of PCR product or displaying apparent homoallelism [13]. With the current methods available, this requires long-range PCR and sequencing of a considerable number of samples that eventually show to be negative for the pathogenic repeat insertion, resulting in a laborious and costly screening assay. To facilitate the diagnostic testing for SCA37, we developed a simple and high-throughput ATTTT RP-PCR that allows a rapid screening of samples.
In routine screening of large populations a simple and inexpensive assay like this ATTTT RP-PCR is highly valuable. We used our control population to assess the highest effort needed in obtaining a conclusive molecular diagnostic. Standard PCR analysis can easily detect pentanucleotide repeat alleles smaller than 30 in DAB1, allowing a conclusive diagnosis in 71% of the samples based on the detection of both alleles (Fig. 3). Larger alleles, however, are not successfully amplified by standard PCR conditions. Therefore, in samples with apparent homoallelism ATTTT RP-PCR proves advantageous by enabling a conclusive negative result in a total of 82% of the samples. On the other hand, this method allows detection of pathogenic alleles with the configuration [(ATTTT)60–79(ATTTC)31–75(ATTTT)58–90] despite the possibility of missing alleles with very different configurations. Samples with a positive ATTTT RP-PCR need to be amplified by long-range PCR followed by sequencing analysis.
Workflow for (ATTTC)n insertion screening. This workflow decreases the number of samples that require long-range PCR followed by Sanger sequencing. First, it is performed a standard PCR, followed by ATTTT RP-PCR in samples wherein only one allele is amplified. The ATTTT RP-PCR distinguishes homoallelism from large allele carriers. In samples with a positive result for the ATTTT RP-PCR long-range PCR and Sanger sequencing are performed to assess the size and composition of large alleles. In the control population studied, 71% of the samples required only standard PCR to complete the negative diagnosis; standard PCR followed by ATTTT RP-PCR in samples with one allele provided a conclusive negative result in 82% of the total samples; the remaining 18% required long-range PCR followed by Sanger sequencing
In conclusion, we suggest that the ATTTT RP-PCR can be used as a rapid screening test in the diagnostic protocol of SCA37 provided this is always followed by long-range PCR amplification and sequencing analysis if a positive ATTTT RP-PCR is detected.
References
Loureiro JR, Oliveira CL, Silveira I. Unstable repeat expansions in neurodegenerative diseases: nucleocytoplasmic transport emerges on the scene. Neurobiol Aging. 2016;39:174–83.
Sequeiros J, Martins S, Silveira I. In Handbook of Clinical Neurology Vol. 103, pp. 227–51, (Netherlands 2012).
Huang M, Verbeek DS. Why do so many genetic insults lead to Purkinje Cell degeneration and spinocerebellar ataxia? Neurosci Lett. (2018) https://doi.org/10.1016/j.neulet.2018.02.004.
Paulson HL, Shakkottai VG, Clark HB, Orr HT. Polyglutamine spinocerebellar ataxias—from genes to potential treatments. Nat Rev Neurosci. 2017;18:613–26.
Coutinho P, Ruano L, Loureiro J, Cruz V, Barros J, Tuna A, et al. Hereditary ataxia and spastic paraplegia in Portugal: a population-based prevalence study. JAMA Neurol. 2013;70:746–55.
Nelson DL, Orr HT, Warren ST. The unstable repeats–three evolving faces of neurological disease. Neuron. 2013;77:825–43.
Garcia-Murias M, Quintans B, Arias M, Seixas AI, Cacheiro P, Tarrio R, et al. ‘Costa da Morte’ ataxia is spinocerebellar ataxia 36: Clinical and genetic characterization. Brain. 2012;135:1423–35.
Alonso I, Jardim L, Artigalas O, Saraiva-Pereira M, Matsuura T, Ashizawa T, et al. Reduced penetrance of intermediate size alleles in spinocerebellar ataxia type 10. Neurology. 2006;66:1602–4.
Silveira I, Alonso I, Guimarães L, Mendonça P, Santos C, Maciel P, et al. High germinal instability of the (CTG)n at the SCA8 locus of both expanded and normal alleles. Am J Hum Genet. 2000;66:830–40.
Silveira I, Miranda C, Guimaraes L, Moreira M, Alonso I, Mendonca P, et al. Trinucleotide repeats in 202 families with ataxia - A small expanded (CAG)(n) allele at the SCA17 locus. Arch Neurol. 2002;59:623–9.
Seixas AI, Vale J, Jorge P, Marques I, Santos R, Alonso I, et al. FXTAS is rare among Portuguese patients with movement disorders: FMR1 premutations may be associated with a wider spectrum of phenotypes. Behav Brain Funct. 2011;7:19.
Sato N, Amino T, Kobayashi K, Asakawa S, Ishiguro T, Tsunemi T, et al. Spinocerebellar ataxia type 31 is associated with “inserted” penta-nucleotide repeats containing (TGGAA)n. Am J Hum Genet. 2009;85:544–57.
Seixas AI, Loureiro JR, Costa C, Ordonez-Ugalde A, Marcelino H, Oliveira CL, et al. A pentanucleotide ATTTC repeat insertion in the non-coding region of DAB1, mapping to SCA37, causes Spinocerebellar Ataxia. Am J Hum Genet. 2017;101:87–103.
Ishikawa K, Dürr A, Klopstock T, Müller S, De Toffol B, Vidailhet M, et al. Pentanucleotide repeats at the spinocerebellar ataxia type 31 (SCA31) locus in Caucasians. Neurology. 2011;77:1853–5.
Cagnoli C, Michielotto C, Matsuura T, Ashizawa T, Margolis RL, Holmes SE, et al. Detection of large pathogenic expansions in FRDA1, SCA10, and SCA12 genes using a simple fluorescent repeat-primed PCR assay. J Mol Diagn. 2004;6:96–100.
Chen L, Hadd A, Sah S, Filipovic-Sadic S, Krosting J, Sekinger E, et al. An information-rich CGG repeat primed PCR that detects the full range of fragile X expanded alleles and minimizes the need for southern blot analysis. J Mol Diagn. 2010;12:589–600.
Acknowledgements
This work was funded by Fundo Europeu de Desenvolvimento Regional-FEDER funds through the COMPETE 2020—Operational Programme for Competitiveness and Internationalisation (POCI), Portugal 2020, and by funding from FCT—Fundação para a Ciência e a Tecnologia/Ministério da Ciência, Tecnologia e Inovação, Portugal, in the framework of the project “Institute for Research and Innovation in Health Sciences” (POCI-01-0145-FEDER-007274); by Grant PTDC/SAU-GMG/098305/2008, from FCT, to I.S. J.R.L. was supported by scholarships from Grant PTDC/GMG-SAU/098305/2008, FCT, PEst-C/SAU/LA0002/2013 and EMBO (ASTF494-2015). C.L.O. was supported by a scholarship from PEst-C/SAU/LA0002/2013. This work was also funded by Norte Portugal Regional Operational Programme (NORTE 2020), under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund (FEDER), Portugal, that supports the Norte-01-0145-FEDER-000008—Porto Neurosciences and Neurologic Disease Research Initiative at I3S.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Loureiro, J.R., Oliveira, C.L., Sequeiros, J. et al. A repeat-primed PCR assay for pentanucleotide repeat alleles in spinocerebellar ataxia type 37. J Hum Genet 63, 981–987 (2018). https://doi.org/10.1038/s10038-018-0474-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-018-0474-3