Abstract
We developed a diagnostic method for repeat expansion diseases using a long-read sequencer to improve currently available, low throughput diagnostic methods. We employed the real-time target enrichment system of the nanopore GridION sequencer using the adaptive sampling option, in which software-based target assignment is available without prior sample enrichment, and built an analysis pipeline that prioritized the disease-causing loci. Twenty-two patients with various neurological and neuromuscular diseases, including 12 with genetically diagnosed repeat expansion diseases and 10 manifesting cerebellar ataxia, but without genetic diagnosis, were analyzed. We first sequenced the 12 molecularly diagnosed patients and accurately confirmed expanded repeats in all with uniform depth of coverage across the loci. Next, we applied our method and a conventional method to 10 molecularly undiagnosed patients. Our method corrected inaccurate diagnoses of two patients by the conventional method. Our method is superior to conventional diagnostic methods in terms of speed, accuracy, and comprehensiveness.
Similar content being viewed by others
Introduction
Tandem repeats (TRs) are a common form of genetic variation in the human genome1. Their expansion may cause disease, usually manifesting a neurological phenotype. To date, approximately 60 TR loci are associated with more than 69 diseases when expanded2. Repeat expansion is responsible for the most common genetic neurology conditions3, whose devastating clinical courses urgently require effective neurotherapies. Very recently, promising neurotherapeutic approaches, such as antisense oligonucleotides4,5, small compounds6, and antibodies7, have been reported for various neurodegenerative diseases. For all these approaches, accurate molecular diagnosis is required.
Molecular diagnosis of repeat expansion disease has been a challenge for neurologists. First, locus heterogeneity is common, necessitating multiple experiments to examine possible loci. Second, TRs are often GC-rich and elongated, making polymerase chain reaction (PCR) difficult. Conventional diagnostic methods are mainly PCR-based, such as flanking PCR and fragment analysis of expanded regions, or repeat-primed PCR (RP-PCR), or depend on southern blotting; therefore, PCR conditions/primers or probes must be specifically set up for each locus. However, this is time consuming and technically difficult, requiring extensive optimization, which can sometimes not be achieved. In practice, several disease loci, but not all, are chosen for diagnosis, sometimes resulting in incomplete screening.
Short-read next-generation sequencing has only contributed to finding new disease-causing repeat expansions to a limited extent; however, long-read sequencing using platforms from Oxford Nanopore Technologies or Pacific Biosciences can potentially cover entire repeat expansions and overcome low-complexity and GC-rich genomic regions. They have boosted recent repeat expansion disease discoveries involving difficult sequences, such as GC-rich repeat motifs8,9 or repeat motifs that are different from those in the reference genome1,10,11,12,13. The currently available methods to capture a region of interest, such as PCR-based enrichment, Cas9-mediated PCR-free enrichment14,15 or Read Until16,17,18, target enrichment software that does not require prior sample preparation, enable long-read sequencing of a targeted region, and they have been recently applied to human genetic research19,20.
Here, we developed a diagnostic method for repeat expansion diseases using long-read sequencing in a clinical setting. We employed real-time target enrichment using a nanopore GridION sequencer with the adaptive sampling option, which is an implemented version of Read Until on GridION, whereby software-based assignment of target regions is available for any repeat expansion disease-associated locus of interest, for up to 1% of a whole-genome region. This enables the GridION sequencer to selectively sequence only the DNA fragments of targets. Furthermore, we built an analysis pipeline that prioritized the disease-causing loci using tandem-genotypes21, a bioinformatic tool that finds changes in the length of TRs from “long” DNA reads aligned to a genome. The pipeline then generates a list of loci with large repeat copy number changes in patients compared with control data in the order of pathogenic possibility. We could easily detect pathogenic repeat expansion without specialist expertise in repeat expansion diseases by following the prioritized list and our data evaluation flowchart. In addition, detailed repeat analyses, such as consensus sequence generation and characterization of interrupting sequence, or methylation analysis, could be performed when needed. From our pilot study of 22 patients with neurological and neuromuscular disorders, we confirm that our diagnostic method is faster, more accurate, and more comprehensive compared with conventional methods.
Results
Evaluation of sequencing quality
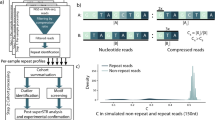
Fifty-nine targeted loci were successfully captured with a mean coverage depth of 24.7 for all 22 patients. Figure 1a shows an example of successful capture at the RFC1 locus. Median depth of coverage across all 59 targeted loci was generally homogenous except for NOTCH2NLC, which might be due to its location in a segmentally duplicated region or its paralogous genes, such as NOTCH2NLA, NOTCH2NLB, and NOTCH2NLR (Supplementary Fig. 1). For detailed evaluation, we plotted the coverage depth of a single run on respective chromosomes for two patients: Patient 8 who had relatively high depth and Patient 5 who had less depth. Relatively homogenous coverage was replicated for on-target reads in these samples. Off-target reads were generally scarce across all chromosomes, although several off-target loci were commonly observed in two patients with relatively high coverage depth (Fig. 1b). Manual inspection revealed that most of these off-target regions did not encompass coding genes, but were located within repetitive regions or at the centromere (data not shown). Even with several highly covered off-targets, targeted regions seemed very accurately enriched overall because the average per-locus coverage depth for on-targets (46.86× for Patient 8 and 12.97× for Patient 5) was roughly 1000 times larger than that for off-targets (0.041× for Patient 8 and 0.0087× for Patient 5) among all selected reads (Fig. 1c).
a The integrative genomics viewer (IGV) depicting the entire RFC1 region successfully captured by adaptive sampling, as an example of successful target enrichment in Patient 9. b Coverage plots for Patient 8 (left) and Patient 5 (right). Upper, middle, and lower panels show coverage plots across whole chromosomes of all reads, on-target reads, and off-target reads, respectively. c All per-locus coverage of “on-target” and coverage per every 5000 bp of “off-target” reads by adaptive sampling were plotted for Patients 8 and 5. The average coverage depths for on- and off-targets are presented in the graph.
Repeat expansions precisely identified in the validation study
All pathogenic repeat expansions irrespective of repeat unit sequence and length were detected in 12 positive controls (Patients 1–12, Table 1, and Fig. 2) and were ranked first in 10 and second in 2 patients by our prioritization workflow (Fig. 3). For validation and discovery studies, loci ranked for possible pathogenicity in each patient are presented in Supplementary Tables 1 and 2, respectively. For patients whose pathogenic locus was ranked second, polymorphic TNRC6A repeat expansion in Patient 3 and heterozygous RFC1 repeat expansion in Patient 4 were ranked as #1. For Patient 3, a TNRC6A repeat expansion was judged as a polymorphism by examining the consensus sequence constructed from our workflow (Supplementary Fig. 2). For Patient 4, an RFC1 repeat expansion was heterozygous; therefore, this patient was considered a carrier by calculating the number of repeat units in the two respective alleles using tandem-genotypes (Supplementary Table 1).
Histograms of tandem-genotypes output and waterfall plots for all positive controls in the validation study are shown. For the histograms, the X-axis indicates copy number change compared with the number of repeat units in the reference human genome: 21 for HTT, 10 for ATXN3, 13 for CACNA1A, 20 for DMPK, 15 for ATXN8OS, 13 for NOTCH2NLC, 20 for PHOX2B, 20 for SAMD12, 11 for RFC1, 13 for BEAN1, 4 for NOP56, and 3 for CSTB. The waterfall plot was generated using either hac mode (Patients 1, 2, 3, 7, 8, 9) or sup mode (Patients 4, 5, 6, 10, 11, 12).
Single targeted long-read sequencing (T-LRS) analysis provided comprehensive results, including whole repeat sequence and expanded repeat length/number of repeat units with its distribution, while conventional methods only provided some of this information for one repeat locus, such as specific repeat unit sequence (RP-PCR), expanded repeat length (southern blotting) or the number of repeat units (fragment length analysis), or only implied that the locus was disease-causing without any detailed data (flanking PCR). Furthermore, the nucleotide-level resolution of T-LRS provided precise information on any interrupting sequences near disease-causing repeats, which can act as disease modifiers or markers. In Patient 1, expanded CAG repeats ended with (CAACAG)1, indicating that this patient will follow the average disease course and severity. Gain or loss of CAACAG sequence influences the age of onset and severity22, which can be a prognostic marker. In Patient 2, CGG, which is associated with intergenerational instability, was confirmed in the mutant allele. In Patient 4, CTA-ATG sequence was inserted at the 5′ end of an expanded repeat. This sequence remained after sequences were basecalled again with super accuracy (sup) mode. Various interrupting sequences, such as CCG, CGG, CAG, and CTC, in the DMPK repeat expansion at the 3′ end and more rarely at the 5′ end have been reported with an estimated frequency of 3–5%, and were associated with a milder phenotype23. The CTA-ATG sequence found in this patient is a previously unreported interrupting sequence with undetermined clinical significance, although we could not exclude the possibility of sequencing error. In Patient 5, pathogenic CTG expansions were observed in both alleles, indicating that this patient had biallelic expansion. The waterfall plot of this patient showed the sequence content of the entire repeat, including a benign CTA repeat, and a disease-causing CTG repeat. The entire repeat sequence content is difficult to clarify using conventional methods, and its pathogenicity was determined based on the total repeat length, which did not exclude the large benign CTA repeat expansion from the pathogenic one. Our T-LRS overcomes this difficulty. In Patient 6, the normal allele had a GGA interruption, which may reduce GGC repeat instability24. In Patient 7, the waterfall plot clearly showed not only repeat length abnormality, but also the detailed sequence content of GCX, where X is A/T/G/C. In Patient 9, homozygous AAGGG repeat expansion was detected. In Patient 10, pathogenic TGGAA, polymorphic TAAAA (common to all ethnic groups), and TAGAA (common in Japanese) repeat unit sequences were all confirmed (Fig. 2).
For Patients 4 and 6 and Individual 1, methylation analysis was performed. Patient 4 was diagnosed with an adult-onset, mild form of myotonic dystrophy having relatively short repeat expansion (approximately 100 repeats). Based on the recent paper reporting that abnormal methylation is mostly observed in the congenital form of myotonic dystrophy, and that patients with larger expanded alleles are more likely to show abnormal methylation25, expanded repeat in this patient may not be hypermethylated. Patient 6 is diagnosed with neuronal intranuclear inclusion disease (NIID); therefore the expanded repeat is expected to be unmethylated. Individual 1 is an unaffected father with an extremely long and hypermethylated repeat expansion in NOTCH2NLC24. As expected, the pathogenic repeat expansions in Patients 4 and 6 were not methylated, while an extremely long repeat expansion in asymptomatic Individual 1 was hypermethylated (Supplementary Fig. 3).
Repeat expansions identified in previously undiagnosed patients
We tested whether we could detect disease-causing repeat expansions in molecularly undiagnosed patients using our method. We examined 10 such patients who had been clinically diagnosed with spinocerebellar ataxia (SCA) (n = 8) or cerebellar ataxia, neuropathy, vestibular areflexia syndrome (CANVAS) (n = 2). Two groups of researchers, who were blind to the results of the respective methods, were assigned to analyze all 10 patients by conventional methods or T-LRS. Both the conventional methods and T-LRS diagnosed 6 out of the 10 patients. Results were different between the conventional methods and T-LRS in two patients (Patients 13 and 17 as described below). For both, the conventional method diagnosis was revised and the T-LRS results were found to be correct (Table 1 and Fig. 4). The results were matched in the remaining patients.
For a–c, the conventional study results are shown on the left and the results of our diagnostic method using GridION are shown on the right. Posi positive control, Nega negative control, NTC no template control, RP-PCR repeat-primed PCR. a Upper left panel shows the flanking PCR result for the CACNA1A locus. Patient 13 and six other patients (1–6) were tested using a 2% agarose gel and only the patient labeled “2” was judged as positive. Upper right panel shows T-LRS results detecting CACNA1A as the rank #1 locus. Lower right panel shows the confirmatory flanking PCR and fragment analysis of the CACNA1A locus. Flanking PCR was evaluated on a 2.5% agarose gel. b Left panel shows flanking PCRs result for the ATXN8OS/ATXN8 locus. Right panel shows the T-LRS result detecting ATXN8OS/ATXN8 as the rank #1 locus but with ambiguous pathogenicity. The rank #2 locus was also rejected for pathogenicity. c Left panel shows an abnormally expanded PCR amplicon in BEAN1, RP-PCR detecting a pathogenic TGGAA repeat, and Sanger sequencing for an SCA31-linked SNP in which one of the patients (Patient 14) did not have this SNP. Right panel shows the T-LRS result detecting the rank #1 locus as BEAN1 and SCA31-linked SNP genotyping shown in the integrative genomics viewer. The result was matched between the two methods, but multiple experiments were needed with the conventional method.
Case studies
Patient 13
CAG expansion in CACNA1A, ranging from 20 to 32 repeats, causes SCA626. The normal repeat unit limit of 18 is close to the abnormal threshold of 20 repeat units; therefore, it may be difficult to diagnose a patient with SCA6 by a conventional method, such as flanking PCR.
For the conventional approach, flanking PCRs targeting 11 different cerebellar ataxia-associated loci were performed (Fig. 5). The first screen was judged negative; therefore, this patient was diagnosed as having no pathogenic repeat expansion. T-LRS was called the rank #1 locus as CACNA1A linked to SCA6, which was compatible with the patient’s phenotype. According to the data evaluation flow, this locus was judged as disease-causing because 1) an abnormal number of repeat units was detected (21 repeat units), 2) it is known to follow autosomal-dominant inheritance, and 3) it is known to have no benign sequence expansion. To confirm this, we performed flanking PCR of the CACNA1A locus again, and carefully checked the size of the PCR amplicon by gel electrophoresis, separating the slightly larger allele from the normal allele of the upper limit size. Fragment analysis confirmed 21 CAG repeat units (Fig. 4a). Retrospectively, this case could be diagnosed with flanking PCR with careful examination, or with fragment analysis without prior screening by flanking PCR. However, this could only be achieved with stringent care or special expertise for repeat expansion diseases and this case showed the clear advantage of T-LRS when detecting small changes in repeat numbers that can be otherwise missed by conventional methods with low resolution.
Square brackets indicate procedures that can be omitted for diagnosis. From this screening workflow, we excluded SCA10, SCA37, and Friedreich ataxia because of their rarity, and SCA36 because of their regional distribution in Japan. DRPLA dentatorubral-pallidoluysian atrophy, CANVAS cerebellar ataxia, neuropathy, vestibular areflexia syndrome.
Patient 17
The bidirectionally transcribed CTG·CAG repeat expansion in ATXN8OS/ATXN8, which leads to mRNA with an expanded CUG repeat and a polyglutamine protein27,28, causes SCA8. Reduced penetrance occurs in SCA8, and a possible modifier of its penetrance is repeat interruption by CCG·CGG29. Normal alleles usually have 15–50 repeats consisting of CTA-CTG·TAG-CAG (preceding polymorphic CTA and subsequent pathogenic CTG) while pathogenic alleles have 71–1300 repeats30.
A large amplicon was detected by SCA8-linked flanking PCR, indicating this patient to be SCA8. T-LRS called the rank #1 locus as ATXN8OS/ATXN8, although it was ambiguous for apparent pathogenicity because its expanded repeat unit number was intermediate (66 repeat units), with a relatively large portion of benign CTA repeat (19 repeat units) compared with previous data showing the number of CTA repeats to be 8–1527, and without CCG interruption. Altogether, this locus was excluded as disease-causing. The rank #2 locus, AXTN7 linked to SCA7, was also rejected for pathogenicity because the repeat expansion did not exceed the manifesting threshold; therefore, this patient had no pathogenic repeat expansion. After comparing the results from the two methods, our final diagnosis was “unlikely pathogenic with intermediate SCA8 repeat expansion”. We cannot completely exclude the possibility of SCA8 with intermediate expansion although repeat length and expanded repeat unit indicated a small chance of disease manifestation. This case showed the advantage of T-LRS when both the number of repeat units and the repeat sequence are important for judging pathogenicity (Fig. 4b).
Patients 14 and 16
SCA31 is relatively common in Japan. A 2.5–3.8-kb-long pentanucleotide repeat expansion consisting of (TGGAA)exp, (TAGAA)exp, (TAAAA)exp, or (TAAAATAGAA)exp in the intronic region of BEAN1 and TK2 is found in patients; however, only TGGAA is linked to disease31,32. There is also a very strongly disease-associated single-nucleotide polymorphism (SNP) in the 5′-untranslated region of PLEKHG432,33, and positive flanking PCR and disease-associated SNPs may confirm SCA31 diagnosis.
SCA31-linked flanking PCR detected a large amplicon, and RP-PCR detected a TGGAA repeat in Patients 14 and 16. Sanger sequencing confirmed that Patient 14 was negative and Patient 16 was positive for the SCA31-linked SNP. T-LRS called BEAN1, linked to SCA31 as the rank #1 locus for both patients, which was judged as pathogenic because abnormal numbers of repeat units were detected (repeat length was 2756 and 2915 bp corresponding to approximately 551 and 583 repeats for Patients 14 and 16, respectively), and the TGGAA sequence (249 and 269 repeats for Patients 14 and 16, respectively) was confirmed in the consensus repeat sequence in both. The SCA31-linked SNP was also targeted; therefore, genotyping information was obtained for both patients by checking the SNP in the integrative genomics viewer without additional experimentation. T-LRS may be advantageous when conventional methods require multiple tests for diagnosis. Additionally, Patient 14 showed that SCA31-linked SNP genotyping cannot be used to exclude a diagnosis of SCA31 (Fig. 4c).
Sequence accuracy
In nanopore sequencing, sequencing accuracy depends on the library preparation kit version used, the Guppy_basecaller version used, and its basecalling model. We used kit 109 and performed basecalling using Guppy v4.3.4, v5.0.11, or v5.1.13 with the basecalling model in high accuracy (hac) mode. According to Oxford Nanopore, raw read accuracy is approximately 95% for Guppy v4.3.4 in hac mode, and 97.8% for Guppy v.5.0.11 or later in hac mode using kit 109 (https://nanoporetech.com/accuracy). When sequences were basecalled again using Guppy v6.0.6 in sup mode, raw read accuracy increased to 98.3%. We therefore performed basecalling again with sup mode for some patients whose waterfall plots had many “other” sequences within the repeat sequence because these were possibly error sequences and might be eliminated with sup mode basecalling. When the waterfall plots of consensus sequences generated in the hac mode were compared with those from the sup mode, the sup mode improved the sequencing accuracy and decreased “other” sequences (Supplementary Fig. 4a). For the AAGGG repeat expansion in RFC1, which causes CANVAS, sequences in Patient 19 were basecalled again with sup mode. However, this resulted in even more “other” sequences in the consensus sequence in a strand-specific manner. Manual inspection detected that most of the “other” sequences were AAGG repeat units (Supplementary Fig. 4b). We previously experienced a similar phenomenon with nanopore sequencing of a CANVAS patient with AAGGG repeat expansion34, so we sequenced this patient (Patient A) using high-fidelity long-read whole-genome sequencing (HiFi LR-WGS) using the PacBio Sequel II system (Pacific Biosciences, Menlo Park, CA, USA). The AAGG repeat observed in T-LRS was mostly absent by PacBio HiFi LR-WGS. Therefore, the “other” sequences in the waterfall plot of Patient 19 are likely to be sequencing/basecalling errors (Supplementary Fig. 4c). Patient 9, another CANVAS patient with AAGGG repeat expansion in RFC1, also showed a “noisy” waterfall plot pattern, similar to that of Patient 19, which may also be because of sequencing errors.
As another way to evaluate sequence accuracy, we correlated the repeat lengths determined by conventional methods and T-LRS using the data from Patients 1, 3, 7, 9, 13, 15, 18, and 19. Significant correlations were observed between repeat lengths determined by conventional methods and T-LRS (P < 0.0001, r2: 0.9822) (Supplementary Fig. 5 and Supplementary Table 3). When validating the correlation for relatively short repeat lengths (up to 150 bp) and large repeat lengths separately, small repeats (n = 9 alleles) showed significant correlation (P < 0.0001, r2: 0.9940), while large repeats (n = 5 alleles) did not reach a statistically significant correlation. This is reasonable because the longer the read, the greater the chance of errors. Alternatively, it may partially result from the limited number of samples used for evaluation. To address this, we increased the number of samples by adding data from previously reported samples34, and reanalyzed the data (n = 16 alleles). This confirmed that large repeats also reached significant correlation (P < 0.0001, r2: 0.9436).
Sensitivity and specificity of T-LRS as a diagnostic method
Our repeat detection workflow outputs a prioritized list of repeat loci in which the loci are displayed in order of importance (i.e., large change in the patient) by comparing them to those from our 27 unaffected controls. This list does not tell the examiner which locus is pathogenic but allows them to judge whether the respective repeat locus has pathogenic repeat expansion or not. If the examiner makes this judgment following the prioritized order from the rank #1 locus, they may easily and rapidly detect pathogenic repeat expansion because pathogenic repeat expansions are all nominated as either rank #1 or #2.
As a diagnostic tool, it is important to provide sensitivity and specificity. However, because this detection flow does not call any locus disease-causing, we could not calculate specificity or sensitivity. As shown in the prioritization list of the top 20 ranked loci for all patients in the validation study (Supplementary Table 1), disease-causing repeat expansion was found in the rank #1 locus in 83.3% (10/12) of the patients, and was found within rank #1 and #2 loci in 100% (12/12) of the patients. This can be a substitute for sensitivity in this detection workflow. No patient was detected in this study with multiple expanded repeats.
As for a substitute for specificity, we checked whether SCA loci other than “true” pathogenic repeat expansions in the six SCA patients (Patients 2, 3, 5, 9, 10, and 11) in the validation study were miscalled as pathogenic repeat expansions. In these patients, other expanded repeats were denied, except for one pathogenic expansion, prior to T-LRS. As shown in Supplementary Table 4, none of the patients had miscalled SCA repeat expansion loci, and only the true pathogenic locus was detected. Thus, the substitute of specificity was 100%.
Time-lag sampling reduces the cost of sequencing
In Japan, it currently costs a minimum of 804 USD to perform one GridION run on one flow cell with two nuclease flushes and all necessary reagents. Conversely, conventional methods cost approximately 6–26 USD depending on how many experiments are needed. To reduce the sequencing cost, we tried “time-lag” sampling to sequence two different samples on one GridION flow cell using nuclease flushes (Fig. 6), which reduced the cost by approximately half (452 USD/sample). We sequenced four patients with CANVAS. Samples 2, 3, and 4 were previously sequenced by HiFi LR-WGS34. Sample 1 was identical to Patient 9. Two samples were serially loaded onto the same flow cell as described in Fig. 6. Mean depth of coverage for the four samples was 15.0× (11.41–16.46×). For all samples, time-lag sampling accurately detected the pathogenic repeat locus (RFC1). The repeat unit sequence [(AAGGG)exp/(AAGGG)exp in samples 1 and 3, (ACAGG)exp/(ACAGG)exp in sample 2, and (AAGGG)exp /(ACAGG)exp in sample 4] and the repeat length were compatible and within 10% of previous results34 (Supplementary Table 5). Regarding the risk of carrying over the previous library to the next sampling, Oxford Nanopore Technologies state that the wash procedure should remove 99.9% of the library, implying that some residual DNA might remain on the flow cell [Nanopore protocol Flow Cell Wash Kit (EXP-WSH004), Version: WFC_9120_v1_revB_08Dec2020]. Assuming 0.1% of the previous library is carried over, approximately 0.015× (15 × 0.001) depth in time-lag sampling might be derived from the previous library. Practically, this can be ignored.
We sequenced four patients with CANVAS, three of whom (Samples 2, 3, and 4) were previously sequenced by HiFi LR-WGS using the PacBio Sequel II system (Pacific Biosciences, Menlo Park, CA, USA)34. Sample 1 was identical to Patient 9. a Time-lag sampling of Sample 1 loading 45 fmol of library on day 1 and Sample 2 loading 55 fmol on day 2 on one GridION flow cell detected pathogenic repeat expansions in both samples. b Time-lag sampling of Sample 3 loading 55 fmol of library on day 1 and Sample 4 loading 55 fmol on day 2 and 15 fmol on day 3 on one GridION flow cell detected pathogenic repeat expansions in both samples. For Sample 4, all remaining library (15 fmol) was loaded again on day 3 because the sequencing output did not reach the satisfactory level at the end of day 2. c Time-lag sampling with Cas9-mediated PCR-free enrichment libraries of Samples 1 and 2. We sequenced Samples 1 and 2 for 7.5 and 24 h, respectively. For Sample 2, there was no AAGGG repeat expansion in its sequencing output, indicating no carryover DNA from Sample 1.
Theoretically, the more depth the sequence output has, the greater the likelihood that the output has carried-over reads from the previous sequencing. To experimentally determine the risk of carryover in our diagnostic system, we performed time-lag sampling with Cas9-mediated PCR-free enrichment of the RFC1 repeat locus to maximize the coverage depth using Samples 1 (repeat unit: AAGGG/AAGGG) and 2 (ACAGG/ACAGG) (Fig. 6). To maximize the depth of Sample 2, Samples 1 and 2 were sequenced for 7.5 and 24 h, respectively. The depths for Samples 1 and 2 were 163.76× and 403.02×, and there were 94 and 375 reads, respectively, which include the entire expanded repeat sequences. For Sample 2, there was no AAGGG repeat expansion in its sequencing output (Fig. 6c). Considering that our method with adaptive sampling would not have such depth, we conclude that we can use time-lag sampling with no practical concern for carrying over the previous sample.
Discussion
Here, we developed a diagnostic method for human repeat expansion diseases using real-time T-LRS on the GridION system with the adaptive sampling option. T-LRS is a time-saving and efficient method that can simultaneously screen multiple disease-associated repeat loci of interest and obtain accurate and comprehensive data on repeat sequence and repeat length/number. T-LRS is also useful for genotyping disease-linked SNPs of interest, which can assist the diagnosis.
We showed that T-LRS has a much higher resolution than conventional methods, enabling finer detection of the repeat expansion and judgment of intermediate expansion that has a low risk of manifesting disease. This advantage improves the sensitivity and specificity of repeat disease diagnostics. Furthermore, recent CAG expansion studies in several diseases, such as SCAs or Huntington’s disease, show that intermediate, non-fully mutated repeat expansion of a locus can be associated with susceptibility to other diseases2,35. This can also happen with amyotrophic lateral sclerosis36,37, frontotemporal dementia38, and Alzheimer's disease39, although there was no such patient in our limited cohort. Therefore, high resolution with multiple locus-targeting T-LRS is preferable.
T-LRS can obtain specific pathogenic repeat unit sequence and interrupting sequence nearby. It has recently been recognized that interrupting sequence can behave as a modifier to affect genetic instability and disease course in either protective or deleterious ways in many repeat expansion disease genes, such as FMR140, ATXN141, HTT22 (with protective effect), and DMPK25,42 (with deleterious or protective effect). Such data would be useful to enable a patient’s disease progression/prognosis to be predicted and to inform a patient’s descendants for family planning. These data can also indicate unknown pathomechanisms or unexpected phenotype-genotype correlations.
One of the merits of our method is the ability to screen as many targets as wanted (up to 1% of the whole genome for efficient enrichment), irrespective of GC content or expanded repeat length, and output is very homogenous with uniform depth of coverage. Second, it can assign target loci without prior experimental preparation, contrary to other target-enriched systems, such as the Cas9-based approach14. We can therefore add a newly identified locus to a target region anytime without any experimental targeting optimization. Third, it can obtain methylation profiles in parallel. Although patients in this study were not suitable for this analysis, we showed that it could be applied to our diagnostic workflow in an ad hoc manner for patients with suspected gene silencing from hypermethylated expanded repeats and adjacent CpG islands, thereby providing additional evidence to support a diagnosis43.
We developed an analysis pipeline that automatically outputs the most likely disease-causing loci with prioritization after sequencing. Diagnosis may be facilitated by following the data evaluation flowchart and may be made without professional experience of repeat expansion diseases.
We also demonstrated a weak point of long-read sequencing, which is decreased raw read accuracy for specific repeat unit sequences, such as AAGGG. Regarding this problem, Tan et al. sequenced telomeres using a Nanopore sequencer with Guppy 5 and Bonito basecalling, and reported extensive basecalling-induced errors at telomere repeats. For example, up to 40–50% of (TTAGGG)n repeats were miscalled as (TTAAAA)n, (TTAAGG)n, (TTAGAG)n, (TTGGGG)n, (CTTCTT)n or (CCCCTGG)n44. They found that miscalling of other types of repeat occurred in a strand-specific manner and that these “other types of repeat” were not observed in the CHM13 reference genome or PacBio HiFi reads, indicating that these “other types of repeat” are artifacts of nanopore sequencing or the basecalling process, rather than biological variation44. The results of Tan et al. seem compatible with our sequencing results of Patient 19, who had an AAGGG repeat expansion.
An important point of this system is that it needs sufficient depth coverage to obtain reliable sequence data that avoids inaccuracies in basecalling and that can separate the two alleles for diagnosing autosomal recessive disease. To estimate the recommended coverage depth for our method, we performed down-sampling of the fastq data from Patients 10 and 18 by various proportions to set the virtual depth of the target region to 4–5×, 7–8×, 10×, 14–15×, and 20–25× (Supplementary Fig. 6). We concluded that to obtain the two separated alleles, a minimum of depth of 10–15× is needed. The amount of output data largely depends on the flow cell quality20; however, one way to increase the coverage depth of a single run is to shear the high-molecular-weight DNA. To detect the expansion of short TRs or minisatellites, the expected size of expansion would be up to 20 kb2; therefore, DNA should be sheared to approximately 40 kb.
Although T-LRS is practical and cost-saving compared with whole-genome sequencing using a long-read sequencer, it is still relatively expensive compared with conventional approaches. However, we believe the various advantages of T-LRS over conventional methods are significant. Furthermore, we propose the time-lag sampling method for reducing its cost by approximately half.
In conclusion, we propose a method for repeat expansion disease diagnostics that can meet the urgent need for rapid, accurate, and comprehensive molecular diagnosis of repeat expansion diseases.
Methods
Patients
Twenty-two patients with various neurological and neuromuscular diseases, including 12 with genetically diagnosed repeat expansion diseases (positive controls) and 10 with clinical diagnosis of SCA or CANVAS, but without genetic diagnosis, were analyzed. Patients 8 and 1245, and Patient 934 were previously described. As a positive control for methylation analysis, unaffected individual 1 who had heterozygous hypermethylated repeat expansion in NOTCH2NLC was analyzed. This individual was recently reported as the asymptomatic father of a patient with NIID24, and is unrelated to Patient 6 with NIID in our cohort. The experimental protocol was approved by the Committee for Ethical Issues at Yokohama City University School of Medicine. Written informed consent was obtained from all individuals. Clinical information was collected from the medical doctors attending the patients. Genomic DNA was extracted using standard methods, from either peripheral blood leukocytes or lymphoblastoid cells established from the patient’s lymphocytes.
Conventional repeat expansion screening
Flanking PCR, Sanger sequencing of PHOX2B repeat region46, and SCA31-linked SNP genotyping which is located in the 5′-untranslated region of PLEKHG4 [rs886041026; PLEKHG4 (NM_001129729.3):c.-16C>T]33, fragment analysis, RP-PCR, and/or southern blotting were applied for genetic diagnoses. Fragment analysis of HTT in Patient 1 and southern blotting of DMPK in Patient 4 were performed at certified clinical laboratories. Our conventional screening workflow for repeat expansion diseases that cause cerebellar ataxia is presented in Fig. 5.
Flanking PCR
Flanking PCR surrounding repeat sequence in genic regions of ATN1 (linked to dentatorubral-pallidoluysian atrophy)47, ATXN1 (linked to SCA1)48, TBP (linked to SCA17)49, PPP2R2B (linked to SCA12)50, ATXN7 (linked to SCA7)51, ATXN8OS/ATXN8 (linked to SCA8)27, ATXN3 (linked to SCA3)52, CACNA1A (linked to SCA6)26, ATXN2 (linked to SCA2)53, BEAN1 (linked to SCA31)32 and RFC1 (linked to CANVAS)54 was performed as previously described, as the first screen of a patient with suspected SCA or/and CANVAS. Primer sequences and PCR conditions are shown in Supplementary Table 6.
Sanger sequencing
Detailed primer sequences and PCR conditions used for Sanger sequencing of PLEKHG4 and PHOX2B are shown in Supplementary Table 6. Sanger sequencing was performed using the Big Dye Terminator cycle sequencing kit (v1.1 or v3.1) (Thermo Fisher Scientific Waltham, MA, USA) on an Applied Biosystems 3500xL Genetic Analyzer (Thermo Fisher Scientific).
Fragment analysis
Fragment analysis for CACNA1A was performed using the same primers and PCR settings as for flanking PCR26 except for the forward primer being 6-carboxyfluorescein (6-FAM)-labeled by Eurofins Genomics (Tokyo, Japan). Primer sequences and PCR conditions are shown in Supplementary Table 6.
RP-PCR
RP-PCR for detecting GGC repeats at NOTCH2NLC (linked to NIID)8, TTTCA repeats at SAMD12 (linked to benign adult familial myoclonic epilepsy)10,45, AAGGG repeats at RFC154, and TGGAA repeats at BEAN1 (linked to SCA31)55 were performed as described8,10,45,54,55. RP-PCR for detecting GGCCTG repeats at NOP56 (linked to SCA36)56 was performed elsewhere. Primer sequences and PCR conditions are shown in Supplementary Table 6. RP-PCR products were resolved and visualized using an Applied Biosystems 3500xL Genetic Analyzer (Thermo Fischer Scientific) and analyzed using GeneMapper software (Thermo Fischer Scientific).
Southern blotting
Patients 8 and 12, on whom southern blotting analysis was performed to detect SAMD12 repeat expansion and CSTB repeat expansion, respectively, were described previously45. Southern blotting for RFC1 intronic repeats was performed as previously reported54. In detail, 3 or 5 µg of genomic DNA was digested overnight with EcoRI-HF (New England Biolabs, Ipswich, MA, USA) to screen for RFC1 repeat expansions. Digested DNA was separated on 0.8% agarose gels in 1.0× Tris-Borate-ethylenediaminetetraacetic acid buffer. The gel was depurinated with 0.25 M HCl for 8 min, and then denaturing DNA was done with 0.5 M NaOH/1.5 M NaCl for 15 min, 2 times. The digested DNA was transferred to positively charged nylon membranes. DNA fragments were fixed using the autocrosslinking mode of the UV Stratalinker 2400 (Stratagene, La Jolla, CA, USA). Prehybridization was performed at 37 °C for 60 min in Digoxigenin (DIG) Easy Hyb buffer (Sigma-Aldrich, St. Louis, MI, USA). DIG-labeled probes were amplified by PCR of a genomic fragment with the forward primer 5’-ATTAGGTGTCTGGTGAGGGC-3’ and the reverse primer 5’-GAAGAATGGCCCCAAAAGCA-3’ (Eurofins Genomics). The PCR products were cloned into a pCR4-TOPO Vector (Invitrogen, Waltham, MA, USA). DIG-labeled RFC1-PCR probe was generated using the PCR DIG Probe Synthesis Kit (Roche, Basel, Switzerland) according to the manufacturer’s instructions. Hybridization was performed at 37 °C overnight in DIG Easy Hyb buffer containing with denatured RFC1-PCR-labeled probe. After hybridization, membranes were washed twice at room temperature in 2× saline-sodium citrate (SSC) containing 0.1% sodium dodecyl sulfate (SDS) for 5 min, followed by two 15-min washes in 0.5× SSC containing 0.1% SDS at 65 °C. The DIG-labeled probes were detected by chemiluminescence, using anti-DIG antibodies conjugated with alkaline phosphatase (Anti-Digoxigenin-AP, Fab fragments, Roche) and its chemiluminescence substrate CDP-star (Roche). Briefly, membranes were blocked for 30 min in 1× blocking solution and then incubated for 30 min in antibody solution (75 mU/mL anti-DIG-AP), followed by two 15-min washes in washing buffer (0.1 M maleic acid, 0.15 M NaCl, 0.3% Tween 20) at room temperature. The signals were visualized on a ChemiDoc Touch (Bio-Rad, Hercules, CA, USA). The membrane was re-probed with custom-made DIG-labeled probes for AAGGG repeat detection ([DIG]-AAGGGAAGGGAAGGGAAGGGAAGGGAAGGGAAGGGAAGGGAAGGG) and ACAGG repeat detection ([DIG]-ACAGGACAGGACAGGACAGGACAGGACAGGACAGGACAGGACAGG) (probes synthesized by Eurofins Genomics).
Real-time T-LRS using adaptive sampling on GridION
Our workflow is presented in Fig. 3. Approximately 2–3 µg of unsheared, purified genomic DNA or DNA sheared to a target size of 40 kb using a Megaruptor 2 (Diagenode, Seraing, Belgium) was used to construct sequencing libraries using the Oxford Nanopore Ligation Sequencing Kit (SQK-LSK109) (Oxford Nanopore Technologies, Oxford, UK) following the manufacturer’s instructions. As an exception to these instructions, enzyme incubation times were doubled as suggested by the manufacturer’s instruction, with the final AMPure purification incubation lasting for 10 min at 37 °C. Approximately 30–50 fmol of library was loaded onto a flow cell (FLO-MIN106D, R9.4.1) on a GridION sequencer (Oxford Nanopore Technologies). Target regions comprising 0.2% of the whole genome were enriched using the adaptive sampling option18 of GridION Mk1 with a bed file assigning 59 loci associated with repeat expansion diseases and each of their surrounding 100 kb regions plus the SCA31-linked SNP in PLEKHG4 and its surrounding 40 kb region (Supplementary Table 7)34. For the technical validation of time-lag sampling, we performed Cas9-mediated PCR-free enrichment for targeting the RFC1 repeat locus in accordance with the manufacturer’s protocols (Nanopore protocol for Cas9-targeted sequencing, ENR_9084_v109_revO_04Dec2018, Oxford Nanopore Technologies) and as previously described24,45,57. The four Alt-R® crRNAs (5′-GACAGUAACUGUACCACAAU-3′ and 5′-ACCACUAGCCAAUGCCUGUU-3′ on the plus strand; 5′-CUAUAUUCGUGGAACUAUCU-3′ and 5′-UAGGACAUUCGGAAAUUCUU-3′ on the minus strand) were mixed and used.
Sequencing was performed in hac mode for approximately 2 to 3 days with one or two additional library loading(s) after nuclease flushing of a flow cell using the Flow Cell Wash Kit (EXP-WSH004) (Oxford Nanopore Technologies). For the time-lag samplings with adaptive sampling target capture, we loaded Sample 1 to the GridION on the first day, and Sample 2 on the second and third days after the nuclease flushes. For the time-lag sampling with Cas9-mediated PCR-free enrichment, we loaded Sample 1 to the GridION on the first day and sequenced for 7.5 h. Sample 2 was loaded after the nuclease flushes and sequenced for 24 h. These procedures are described in Fig. 6.
Data analysis
We built a data analysis pipeline using the analysis tools mentioned below, and evaluated the output data from the pipeline, following the data evaluation workflow presented in Fig. 3. Sequences were basecalled using Guppy v4.3.4, v5.0.11, or v5.1.13 in hac mode during the run on the GridION. According to Oxford Nanopore, sup mode increases the raw read accuracy; therefore, sequences were basecalled again using Guppy v6.0.6 in sup mode for Patients 4, 5, 6, 10, 11, 12, 14, and 16. They were then aligned to GRCh38 using either minimap2 v2.14 (https://github.com/lh3/minimap2) or LAST 1132 (https://gitlab.com/mcfrith/last). Depth of coverage was calculated using mosdepth v0.3.1 (https://github.com/brentp/mosdepth). Median and range of coverage depth across all 59 loci among 22 patients were calculated (Supplementary Fig. 1). Tandem-genotypes v1.3.0, v1.8.2, or v1.9.0 (https://github.com/mcfrith/tandem-genotypes) was used to find changes in the length of TRs of selected loci. Histograms of the output were drawn using tandem-genotypes-plot command, where x-axis, “copy number change”, indicates difference in the number of repeat units relative to the reference human genome, while red and blue bars indicate the forward- and the reverse-strand reads. Crude allele prediction was performed with the tandem-genotypes -o2 option, and they are shown as consensus differences in the numbers of repeat units compared with reference human genome sequence for each of the two alleles (Supplementary Tables 1 and 2). For prioritization of pathogenic repeat expansion within detected repeats, tandem-genotypes-join was performed against data from 27 controls sequenced using PromethION (Oxford Nanopore Technologies). These 27 controls were healthy Japanese individuals with no neurodegenerative disorders. For the 46 out of 59 targeted loci that are especially relevant to known repeat expansion diseases that follow Mendelian inheritance, the distribution of the number of repeat units in 54 alleles from our 27 control samples is plotted in Supplementary Fig. 7. Consensus sequences were constructed using lamassemble 1.4.2 (https://gitlab.com/mcfrith/lamassemble) or the tandem-genotypes merge option implemented in version 1.8.0 or later. When pathogenic repeat expansion was detected, we proceeded to the additional analyses (in an ad hoc manner), including detailed repeat analysis and methylation analysis. Detailed repeat analyses were performed using RepeatAnalysisTools, including the generation of waterfall-style plots from a consensus fasta file of the region of interest (waterfall plot) or coverage plots of the mapped reads (https://github.com/PacificBiosciences/apps-scripts/tree/master/RepeatAnalysisTools). When abnormal methylation was suspected, methylation analysis was performed as described previously24, except that we used Guppy v6.0.6 basecaller to detect 5-methylcytosine with the configuration file named “dna_r9.4.1_450bps_modbases_5mc_hac.cfg”, and we employed default settings for methylation detection and in silico bisulfite-like conversion.
To evaluate the accuracy and efficiency of target enrichment, we generated quality-filter passed fastq files, excluding reads of 1000 bp or less, which are judged by GridION to be “out of target” reads and are quit from sequencing on the adaptive sampling principle, using Filtlong v0.2.1 (https://github.com/rrwick/Filtlong). Then, we calculated the depth of coverage against both on and off-target regions among all selected reads using mosdepth. For on-targets, the average “per-locus depth of coverage” was calculated for all loci. For off-targets, bam files, including reads outside the targeted regions assigned by the bed file, were generated using samtools view -L option, and the coverage depth for every bin of 5000 bp was calculated using mosdepth. The averages of on- and off-target coverage depth data were compared to evaluate enrichment efficiency.
For down-sampling of the fastq data, we used seqkit v0.11.0 with the “sample” command by assigning the various proportions of down-sampling with -p option to set the virtual depth of the target region at 4–5×, 7–8×, 10×, 14–15× and 20–25× (https://bioinf.shenwei.me/seqkit/usage/#sample). We performed the down-sampling on fastq data from Patients 10 and 18.
Correlation analysis between conventional methods and T-LRS on GridION
The repeat length detected by any of the conventional methods (fragment analysis, Sanger sequencing, or southern blotting) and T-LRS were compared, and the correlation between the two for Patients 1, 3, 7, 9, 13, 15, 18, and 19 was analyzed statistically by the linear regression model using GraphPad Prism v8.0.2 (GraphPad Software, San Diego, CA USA). A P value <0.05 was considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The datasets for this article are not publicly available because of concerns regarding patients’ anonymity (our data are considered as a personal identifier since it contains more than nine loci of 4-bp short tandem repeats (STRs), which is defined as a personal identifier under the Japanese law concerning protection of personal information). Requests to access the datasets from qualified researchers should be directed to the corresponding author. There are restrictions on a qualified researcher accessing the data (non-commercial use only and requiring a Data Usage Agreement).
Code availability
All computational tools used in this study are available as open-source software, and their download links are presented in Methods. Software used are: Guppy v4.3.4, v5.0.11, v5.1.13, or v6.0.6 (https://nanoporetech.com/), minimap2 v2.14 or v2.23 (https://github.com/lh3/minimap2), LAST 1132 (https://gitlab.com/mcfrith/last), mosdepth v0.3.1 (https://github.com/brentp/mosdepth), Tandem-genotypes v1.3.0, v1.8.2, or v1.9.0 (https://github.com/mcfrith/tandem-genotypes), lamassemble 1.4.2 (https://gitlab.com/mcfrith/lamassemble), RepeatAnalysisTools (https://github.com/PacificBiosciences/apps-scripts/tree/master/RepeatAnalysisTools), methyl-stat that is a collection of command-line tools for analyzing methylation-aware oxford nanopore sequencing data including methyl-stat, methylcall, and ont2bisul (https://github.com/bitsyamagu/methyl-stat), Filtlong v0.2.1 (https://github.com/rrwick/Filtlong), seqkit v0.11.0 (https://github.com/shenwei356/seqkit), and samtools v1.3.1 or 1.9 (https://github.com/samtools/samtools). As a specific parameter, we used -r 50000 --end-bonus 10000 –no-end-fit for better mapping of long repeat regions using minimap2 as described previously19.
References
Depienne, C. & Mandel, J. L. 30 years of repeat expansion disorders: what have we learned and what are the remaining challenges? Am. J. Hum. Genet. 108, 764–785 (2021).
Gall-Duncan, T., Sato, N., Yuen, R. K. C. & Pearson, C. E. Advancing genomic technologies and clinical awareness accelerates discovery of disease-associated tandem repeat sequences. Genome Res. 32, 1–27 (2022).
Lockhart, P. J. Advancing the diagnosis of repeat expansion disorders. Lancet Neurol. 21, 205–207 (2022).
Tran, H. et al. Suppression of mutant C9orf72 expression by a potent mixed backbone antisense oligonucleotide. Nat. Med. 28, 117–124 (2022).
Ellerby, L. M. Repeat expansion disorders: mechanisms and therapeutics. Neurotherapeutics 16, 924–927 (2019).
Nakamori, M. et al. A slipped-CAG DNA-binding small molecule induces trinucleotide-repeat contractions in vivo. Nat. Genet. 52, 146–159 (2020).
Nguyen, L. et al. Antibody therapy targeting RAN proteins rescues C9 ALS/FTD phenotypes in C9orf72 mouse model. Neuron 105, 645–662.e611 (2020).
Sone, J. et al. Long-read sequencing identifies GGC repeat expansions in NOTCH2NLC associated with neuronal intranuclear inclusion disease. Nat. Genet. 51, 1215–1221 (2019).
Ishiura, H. et al. Noncoding CGG repeat expansions in neuronal intranuclear inclusion disease, oculopharyngodistal myopathy and an overlapping disease. Nat. Genet. 51, 1222–1232 (2019).
Ishiura, H. et al. Expansions of intronic TTTCA and TTTTA repeats in benign adult familial myoclonic epilepsy. Nat. Genet. 50, 581–590 (2018).
Florian, R. T. et al. Unstable TTTTA/TTTCA expansions in MARCH6 are associated with Familial Adult Myoclonic Epilepsy type 3. Nat. Commun. 10, 4919 (2019).
Yeetong, P. et al. TTTCA repeat insertions in an intron of YEATS2 in benign adult familial myoclonic epilepsy type 4. Brain 142, 3360–3366 (2019).
Corbett, M. A. et al. Intronic ATTTC repeat expansions in STARD7 in familial adult myoclonic epilepsy linked to chromosome 2. Nat. Commun. 10, 4920 (2019).
Giesselmann, P. et al. Analysis of short tandem repeat expansions and their methylation state with nanopore sequencing. Nat. Biotechnol. 37, 1478–1481 (2019).
Gilpatrick, T. et al. Targeted nanopore sequencing with Cas9-guided adapter ligation. Nat. Biotechnol. 38, 433–438 (2020).
Payne, A. et al. Readfish enables targeted nanopore sequencing of gigabase-sized genomes. Nat. Biotechnol. 39, 442–450 (2021).
Kovaka, S., Fan, Y., Ni, B., Timp, W. & Schatz, M. C. Targeted nanopore sequencing by real-time mapping of raw electrical signal with UNCALLED. Nat. Biotechnol. 39, 431–441 (2021).
Loose, M., Malla, S. & Stout, M. Real-time selective sequencing using nanopore technology. Nat. Methods 13, 751–754 (2016).
Miller, D. E. et al. Targeted long-read sequencing identifies missing disease-causing variation. Am. J. Hum. Genet. 108, 1436–1449 (2021).
Stevanovski, I. et al. Comprehensive genetic diagnosis of tandem repeat expansion disorders with programmable targeted nanopore sequencing. Sci. Adv. 8, eabm5386 (2022).
Mitsuhashi, S. et al. Tandem-genotypes: robust detection of tandem repeat expansions from long DNA reads. Genome Biol. 20, 58 (2019).
Wright, G. E. B. et al. Length of uninterrupted CAG, independent of polyglutamine size, results in increased somatic instability, hastening onset of Huntington disease. Am. J. Hum. Genet. 104, 1116–1126 (2019).
Peric, S., Pesovic, J., Savic-Pavicevic, D., Rakocevic Stojanovic, V. & Meola, G. Molecular and clinical implications of variant repeats in myotonic dystrophy type 1. Int. J. Mol. Sci. 23, 354 (2021).
Fukuda, H. et al. Father-to-offspring transmission of extremely long NOTCH2NLC repeat expansions with contractions: genetic and epigenetic profiling with long-read sequencing. Clin. Epigenetics 13, 204 (2021).
Morales, F. et al. Myotonic dystrophy type 1 (DM1) clinical subtypes and CTCF site methylation status flanking the CTG expansion are mutant allele length-dependent. Hum. Mol. Genet. 31, 262–274 (2021).
Zhuchenko, O. et al. Autosomal dominant cerebellar ataxia (SCA6) associated with small polyglutamine expansions in the alpha 1A-voltage-dependent calcium channel. Nat. Genet. 15, 62–69 (1997).
Koob, M. D. et al. An untranslated CTG expansion causes a novel form of spinocerebellar ataxia (SCA8). Nat. Genet. 21, 379–384 (1999).
Moseley, M. L. et al. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat. Genet. 38, 758–769 (2006).
Perez, B. A. et al. CCG*CGG interruptions in high-penetrance SCA8 families increase RAN translation and protein toxicity. EMBO Mol. Med. 13, e14095 (2021).
Hu, Y. et al. Sequence configuration of spinocerebellar ataxia type 8 repeat expansions in a Japanese cohort of 797 ataxia subjects. J. Neurol. Sci. 382, 87–90 (2017).
Ishiguro, T., Nagai, Y. & Ishikawa, K. Insight into spinocerebellar ataxia type 31 (SCA31) from Drosophila model. Front. Neurosci. 15, 648133 (2021).
Sato, N. et al. Spinocerebellar ataxia type 31 is associated with “inserted” penta-nucleotide repeats containing (TGGAA)n. Am. J. Hum. Genet. 85, 544–557 (2009).
Ishikawa, K. et al. An autosomal dominant cerebellar ataxia linked to chromosome 16q22.1 is associated with a single-nucleotide substitution in the 5’ untranslated region of the gene encoding a protein with spectrin repeat and Rho guanine-nucleotide exchange-factor domains. Am. J. Hum. Genet. 77, 280–296 (2005).
Miyatake, S. et al. Repeat conformation heterogeneity in cerebellar ataxia, neuropathy, vestibular areflexia syndrome. Brain 145, 1139–1150 (2022).
Tezenas du Montcel, S. et al. Modulation of the age at onset in spinocerebellar ataxia by CAG tracts in various genes. Brain 137, 2444–2455 (2014).
Elden, A. C. et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 466, 1069–1075 (2010).
Conforti, F. L. et al. Ataxin-1 and ataxin-2 intermediate-length PolyQ expansions in amyotrophic lateral sclerosis. Neurology 79, 2315–2320 (2012).
Fournier, C. et al. Interrupted CAG expansions in ATXN2 gene expand the genetic spectrum of frontotemporal dementias. Acta Neuropathol. Commun. 6, 41 (2018).
Rosas, I. et al. Role for ATXN1, ATXN2, and HTT intermediate repeats in frontotemporal dementia and Alzheimer’s disease. Neurobiol. Aging 87, 139.e131–139.e137 (2020).
Eichler, E. E. et al. Length of uninterrupted CGG repeats determines instability in the FMR1 gene. Nat. Genet. 8, 88–94 (1994).
Chong, S. S. et al. Gametic and somatic tissue-specific heterogeneity of the expanded SCA1 CAG repeat in spinocerebellar ataxia type 1. Nat. Genet. 10, 344–350 (1995).
Braida, C. et al. Variant CCG and GGC repeats within the CTG expansion dramatically modify mutational dynamics and likely contribute toward unusual symptoms in some myotonic dystrophy type 1 patients. Hum. Mol. Genet 19, 1399–1412 (2010).
Chintalaphani, S. R., Pineda, S. S., Deveson, I. W. & Kumar, K. R. An update on the neurological short tandem repeat expansion disorders and the emergence of long-read sequencing diagnostics. Acta Neuropathol. Commun. 9, 98 (2021).
Tan, K., Slevin, M., Meyerson, M. & Li, H. Identifying and correcting repeat-calling errors in nanopore sequencing of telomeres. Genome Biol. 23, 180 (2022).
Mizuguchi, T. et al. Complete sequencing of expanded SAMD12 repeats by long-read sequencing and Cas9-mediated enrichment. Brain 144, 1103–1117 (2021).
Matera, I. et al. PHOX2B mutations and polyalanine expansions correlate with the severity of the respiratory phenotype and associated symptoms in both congenital and late onset Central Hypoventilation syndrome. J. Med. Genet. 41, 373–380 (2004).
Koide, R. et al. Unstable expansion of CAG repeat in hereditary dentatorubral-pallidoluysian atrophy (DRPLA). Nat. Genet. 6, 9–13 (1994).
Orr, H. T. et al. Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nat. Genet. 4, 221–226 (1993).
Nakamura, K. et al. SCA17, a novel autosomal dominant cerebellar ataxia caused by an expanded polyglutamine in TATA-binding protein. Hum. Mol. Genet 10, 1441–1448 (2001).
Holmes, S. E. et al. Expansion of a novel CAG trinucleotide repeat in the 5’ region of PPP2R2B is associated with SCA12. Nat. Genet. 23, 391–392 (1999).
David, G. et al. Cloning of the SCA7 gene reveals a highly unstable CAG repeat expansion. Nat. Genet. 17, 65–70 (1997).
Kawaguchi, Y. et al. CAG expansions in a novel gene for Machado-Joseph disease at chromosome 14q32.1. Nat. Genet. 8, 221–228 (1994).
Pulst, S. M. et al. Moderate expansion of a normally biallelic trinucleotide repeat in spinocerebellar ataxia type 2. Nat. Genet. 14, 269–276 (1996).
Cortese, A. et al. Biallelic expansion of an intronic repeat in RFC1 is a common cause of late-onset ataxia. Nat. Genet. 51, 649–658 (2019).
Ishige, T. et al. Pentanucleotide repeat-primed PCR for genetic diagnosis of spinocerebellar ataxia type 31. J. Hum. Genet. 57, 807–808 (2012).
Kobayashi, H. et al. Expansion of intronic GGCCTG hexanucleotide repeat in NOP56 causes SCA36, a type of spinocerebellar ataxia accompanied by motor neuron involvement. Am. J. Hum. Genet. 89, 121–130 (2011).
Nakamura, H. et al. Long-read sequencing identifies the pathogenic nucleotide repeat expansion in RFC1 in a Japanese case of CANVAS. J. Hum. Genet. 65, 475–480 (2020).
Acknowledgements
We thank all participants for their cooperation in this research. We are grateful to K. Takabe, Watanabe, and S. Sugimoto from the Department of Human Genetics, Yokohama City University Graduate School of Medicine, and Dr Martin Frith from the Artificial Intelligence Research Center, National Institute of Advanced Industrial Science and Technology (AIST), for their technical assistance. We also thank Jeremy Allen, PhD, from Edanz (https://jp.edanz.com/ac) for editing a draft of this manuscript. This work was supported by the Japan Agency for Medical Research and Development (AMED) under grant numbers JP21ek0109486, JP21ek0109549, JP21cm0106503, and JP21ek0109493 (N.M.); JSPS KAKENHI under grant numbers JP20K07907 (S.M.), JP18K07503 (H.D.), JP21K07869 (E.K.), JP20K16932 (K.Ham.), JP20K17936 (A.F.), JP21K15907 (Y.U.), JP20K17428 (N.T.), and JP20K08164 (T.Miz.); the Takeda Science Foundation (H.D., T.Miz., and N.M.), and a Grant for Strategic Research Promotion from Yokohama City University (F.T.: #SK2804).
Author information
Authors and Affiliations
Contributions
S.M. and N.M. contributed to the conception and design of the study. S.M., E.K., A.F., H.D., K.Ham., M.O., T.W., N.U., H.K., G.M., A.M., M.K., K.O., R.K., A.Su., A.Sa., T.Miy., M.S., Y.U., N.T., H.H., K.M., K.Hay., Y.S., H.A., K.Y., F.T., T.Miz., and N.M. contributed to the acquisition and analysis of data. S.M., E.K., and N.M. contributed to drafting the text. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Miyatake, S., Koshimizu, E., Fujita, A. et al. Rapid and comprehensive diagnostic method for repeat expansion diseases using nanopore sequencing. npj Genom. Med. 7, 62 (2022). https://doi.org/10.1038/s41525-022-00331-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41525-022-00331-y
This article is cited by
-
Sequence composition changes in short tandem repeats: heterogeneity, detection, mechanisms and clinical implications
Nature Reviews Genetics (2024)
-
Prevalence of repeat expansions causing autosomal dominant spinocerebellar ataxias in Hokkaido, the northernmost island of Japan
Journal of Human Genetics (2024)
-
Next-generation sequencing and bioinformatics in rare movement disorders
Nature Reviews Neurology (2024)
-
The expanding diagnostic toolbox for rare genetic diseases
Nature Reviews Genetics (2024)
-
Sequencing and characterizing short tandem repeats in the human genome
Nature Reviews Genetics (2024)