Abstract
We present new data and analysis on the genetic variation of contemporary inhabitants of central Argentina, including a total of 812 unrelated individuals from 20 populations. Our goal was to bring new elements for understanding micro-evolutionary and historical processes that generated the genetic diversity of the region, using molecular markers of uniparental inheritance (mitochondrial DNA and Y chromosome). Almost 76% of the individuals show mitochondrial lineages of American origin. The Native American haplogroups predominate in all surveyed localities, except in one. The larger presence of Eurasian maternal lineages were observed in the plains (Pampas) of the southeast, whereas the African lineages are more frequent in northern Córdoba. On the other hand, the analysis of 258 male samples reveals that 92% of them present Eurasian paternal lineages, 7% carry Native American haplogroups, and only 1% of the males show African lineages. The maternal lineages have high genetic diversity homogeneously distributed throughout central Argentina, probably as result of a recent common origin and sustained gene flow. Migratory events that occurred in colonial and recent times should have contributed to hiding any traces of differentiation that might have existed in the past. The analysis of paternal lineages showed also homogeneous distribution of the variation together with a drastic reduction of the native male population.
Similar content being viewed by others
Introduction
The central region of Argentina has been inhabited by humans since the late Pleistocene [1,2,3]. Archaeologically known as Sierras Centrales [4], it is possible to recognize at least two different stages in the human occupation of the region, associated with two different ways of life: a hunter-gathered subsistence system, from the first settlements (around 11000 yrs BP) until the adoption of horticulture, llama breeding, and sedentarism in pit-houses (about 1500 yrs BP). The populations that inhabited the mountains chains of Córdoba and San Luis provinces were denominated “Comechingones”, whereas the inhabitants of northeastern of Cordoba and south of Santiago of the Estero were all called “Sanavirones” [5]. Unfortunately, these homogenizing designations hid temporal, biological [6,7,8], and cultural variation that existed in the region [9,10,11,12].
In the five centuries elapsed from the first occupation of the Argentine territory by the Kingdom of Spain to the present, two main stages are distinguished in the migration process. The first stage corresponds to the expansion of the territory under Spanish domination [13], and is characterized by the arrival of Europeans, and Africans as slave labor force. The second stage, between the second half of the nineteenth century and early twentieth century, is characterized by a large migratory wave of European and Middle Eastern peoples as part of a process of economic internationalization [14]. Different demographic processes characterized these two periods. In colonial times (from the sixteenth century until the early nineteenth), when Spaniards and enslaved African settled the territory, the population was predominantly rural and concentrated mainly on Córdoba city and the Sierras region, which was also the principal place of the ancient indigenous settlement and the earliest Spanish occupation. On the other hand, the Pampas plains of the Southeast, with lower indigenous population density, was more recently occupied by the migrants. Around mid-nineteenth century began a shift in the valuation of the Pampas soils in the context of an emerging agricultural and cattle economy [15]. This change occurs shortly before the massive influx of European immigrants (1880–1930), who settled mainly in large cities and in the Pampas and Patagonian territories, as part of the State policy of colonization and “civilization” of the country. This migration wave is known as “the European immigration flood”, and led to the settlement of about 3 million Europeans. The National Census of 1914 indicated 20%, 9%, and 4% of foreigners residing in Cordoba, San Luis, and Santiago del Estero, respectively. Most of these migrants came from Spain and Italy and, in a lesser degree, from Germany, Russia, and France.
Evidence obtained by our research group and colleagues shows that after five centuries of colonization and cultural exchange, the gene pool of indigenous populations is still present in the contemporary population of central Argentina [16,17,18]. This is particularly true for maternal lineages, whereas the survival of native paternal lineages is very low [19]. This discrepancy is common throughout Latin America, reflecting a pattern of directional mating involving European men and Native American and African women [20, 21]. Based on the analysis of 10 autosomal ancestry informative markers (AIMs), García et al. [22]. studied the admixture patterns of the same populations and found that the main parental contributions to the gene pool of Córdoba and San Luis provinces corresponds to Europeans (55% and 58%, respectively) and Native Americans (44% and 40%), whereas the African component is very low, being within the range of statistical error (1.3% and 1.5%).
In this article, we present new data and analysis of the genetic variation of the contemporary inhabitants of Central Argentina, including 20 population samples across three provinces (Córdoba, San Luis, and Santiago del Estero), based on molecular markers of uniparental inheritance. Our goal is to bring new elements for understanding micro-evolutionary and historical processes that generated the current genetic diversity of the region.
Materials and methods
Population samples
A total of 812 buccal swab samples were obtained from unrelated individuals, from 14 villages of Córdoba (N = 606), four of San Luis (N = 121), and two of Santiago del Estero (N = 85) provinces. Figure 1a shows the map of the area with the sampling places. The studied region is located in the southern part of the Sierras Pampeanas of Argentina, at latitudes from 28.5°S to 34.5°S and longitudes between 61.8°W and 66°W, covering mountain ranges (Sierras) and plains (Pampas) of Córdoba, northern San Luis, and southern Santiago del Estero provinces. Some of the samples have been previously analyzed, specifically, the samples from Santiago del Estero (N = 85) were reported by Pauro et al. [18], whereas a subset of samples belonging to haplogroup D1 from the three provinces (N = 71) were analyzed in the article of García et al. [23]. The sample collection was carried out in public hospitals and community health centers. All the participants were informed of the objectives of this nonprofit scientific investigation and signed a written informed consent, which was approved by the Ethical Committees of the CEMIC (Comité de Ética en Investigación, Centro de Educación Médica e Investigaciones Clínicas “Norberto Quirno”). Information gathered in the field regarding the birthplaces of subjects and their parents indicates that the inhabitants of the studied populations present low to moderate mobility and were limited to their region of origin for the last two or three generation in most of the cases, presenting the rest of the individuals extracontinental ancestors. The exception is Cordoba city, where a more diverse population was found, which can be explained by the industrialization process of the last 50 years. See further information in Material and methods (at the end of population sample section). The sample sizes for each locality are presented in Table 1.
Geographic location of populations. a Sampling locations of the central Argentina populations. References for populations and their abbreviated names are from Table 1. b Map showing the geographical location from populations of different regions of the Southern Cone included in the maternal lineages analysis. References for populations and their abbreviated names are from Table 2
In addition, we included in the analysis mitochondrial DNA (mtDNA) HVRI sequences from 44 Southern Cone populations, comprising a total of 2511 individuals (Fig. 1b) obtained from published sources (Table 2). Comparisons with other populations from different regions of the Southern Cone were carried out with the aim of analyzing the genetic diversity of central Argentina in a larger geographical scale. We included data from populations representing the major geographical regions of the Southern Cone (Table 2), as Gran Chaco, subtropical forests, Central Andes, Patagonia, and Tierra del Fuego, as well as historically distinctive regions, such as Cuyo (western Argentina), and NWA (northwestern Argentina). Finally, 19 samples from Argentina, Chile, and Paraguay, and samples from other regions than Central Argentina drawn from Bailliet et al. [24] were included for Y chromosome comparisons.
Laboratory methods
Genomic DNA was extracted from cheek swabs using the Accuprep Genomic DNA Purification Kit (GenBiotech). All samples were assigned to the major haplogroups by restriction fragment length polymorphism (PCR-RFLP) [25] or amplified product-length polymorphism multiplex (PCR-APLP) [26]. mtDNA non-coding control regions (HVRI and HVRII) were amplified using the following specific primers F15878 (5´-AAATGGGCCTGTCCTTGTAG-3´), F16475 (5´-TAGCTAAAGTGAACTGTATCC-3´), and R633 (5´-GCATGTGTAATCTTACTAAGAG-3´). The amplification reactions were performed in a Biometra T-Personal thermocycler in a volume of 50 ul under the following conditions: an initial denaturation step of 94 °C for 5 min, followed by 40 cycles with temperatures of 94 °C for 1 min, 52 °C for 1 min, and 72 °C for 1.5 min, with a final extension step of 5 min at 72 °C. Verification and quality control of PCR amplification was performed by 2% agarose gel electrophoresis, stained with GelStar (Lonza) and visualized with ultraviolet light. The amplified products were sent to Macrogen Inc. (Seoul, Korea) for purification and automatic sequencing. In those cases where the sequence was interrupted by the presence of polyC tract at positions 16,184–16,193 and/or 303–309/303–315, the following additional internal primers were used: F16190 (5´-CCCCATGCTTACAAGCAAGT-3´), R16410 (5´-GAGGATGGTGGTCAAGGAC-3´), and F314 (5´-CCGCTTCTGGCCACAGCACT-3´). The haplotypes and the polymorphic sites are listed in Supplementary Table S1, where the final reading range for each sample is detailed.
Sequences were corrected manually and then aligned and compared with the revised Cambridge Reference Sequence [27] using the program Sequencher (Sequencher® version 5.2 sequence analysis software, Gene Codes Corporation, Ann Arbor, MI USA; http://www.genecodes.com). To define the haplogroups, we used the HaploGrep2 [28] and Phylotree Build 17 [29]. Membership of maternal lineages into the most derived clades was performed by critically employing Haplogrep. Initial review of assignations allowed us to note the presence of rather unusual lineages in Central Argentina given the expectations derived from the well-known phylogeographical structure of sub-haplogroups on continental scale. As most of these assignations were based on the presence of single, medium-to-fast evolving sites in control region like 146, 151, 16,192 etc., we decided to retain only those clades whose definitions were inequivocal and/or whose presence in the area had been previously informed based on mitogenomic information. Furthermore, we interpreted all the B4b lineages as B2, as B2 is only B4b branch known to be present in the Americas and easily distinguishable from its sister branch B4b1 from Asia. Finally, the haplogroup assignment was confirmed using the software MitoTool [30].
Two hundred fifty-eight male samples were analyzed with eight biallelic markers located in the nonrecombinant region of the Y chromosome. These polymorphisms were defined by the presence or absence of the characteristic mutation for the following markers: M3 [31], M207, M173 [32], M168, M9 [33], M89 [34], M2 [35], and YAP [36]. The polymorphisms were analyzed using two different protocols: (1) for RFLP-PCR following standard conditions with minor modifications and then digested with restriction enzymes (except for the insertion YAP), and (2) for APLP-PCR following the protocol of Jurado et al. [37], based in the specific alellic co-amplification. The amplification strategy was carried out following a hierarchical amplification protocol [38], which means that each subject was not genotyped for all markers. The genotyped samples were assigned to haplogroups, useful for identifying geographic origins [39, 40].
Data analysis
Population structure, both at regional and continental levels, was investigated by mean of the analysis of molecular variance (AMOVA) [41]. Different estimators were employed to investigate genetic diversity in the studied population: haplotype number (k), polymorphic sites (s), haplotype diversity (Ĥ), nucleotide diversity (π), and tests of neutral selection such as Tajima´D (D) and Fu´s test (FS). These analyses were performed using the program Arlequin, version 3.5 [42]. This program was also used to calculate pairwise genetic distances, Kimura 2-parameters [43] (K2P), based on mtDNA HVRI sequences. From the distance matrix, we performed discriminant analysis (DA) between geographic groups of populations (as presented in Table 2) using principal coordinates analysis [44] as a prior step, transforming the K2P matrix into vectors representing the populations onto a multivariate space. Without implying a loss of genetic information, this transformation allows DA to be applied and attempts to summarize the genetic differentiation between groups. This method also allows for a probabilistic assignment of individuals (in this case populations) to each group, as in Bayesian clustering methods [45]. This analysis was performed excluding variants 16182C–16183C, the variation in number of cytosines around nps 309–315, and indels 514–524.
Spatial analysis
Isofrequency maps were generated for a visual analysis of the distribution of continental mitochondrial lineages in Central Argentina using the program Surfer 7 (Golden Software, Golden, CO), following the Kriging procedure. At continental level, we computed a geographic distance matrix as spherical distances in kilometers between pairs of populations. This distance matrix was later used to test for any association between K2p distances and geographic distances through matrix correlation analysis. Statistical significance was evaluated using the Mantel test [46]. We also performed the global spatial autocorrelation method of Smouse and Peakall [47]. For this purpose, we constructed a correlogram representing the correlation between the K2P and the geographic distances matrices across 10 distance classes. The autocorrelation coefficient r provides a measure of the genetic similarity between pairs of populations whose geographic separation falls within the specified distance class. After that, the Bearing analysis was employed for determining the spatial directionality of the mtDNA variation. The Bearing analysis [48] is a method of determining the direction of greatest correlation between data distance and geographic distance. The procedure starts with two matrices: a data distance matrix V and a geographic distance matrix D. The correlation between V and D was calculated via a Mantel test for 36 fixed bearings, each differing by 5 degrees. For spatial analysis techniques, we used the program Passage 2.0 [49].
For the analysis of population relationships based on Y chromosome haplogroup frequencies, we calculated chord distances between pairs of populations. From the distance matrix, a genetic map was constructed by means of multiple dimensional scaling [50]. These analyses were carried out using the program PAST 3.0 [51].
Results
Analysis of maternal lineages
Distribution of mitochondrial lineages according to continental origin
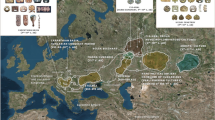
Out of the 812 samples analyzed in this study, 616 (76%) have mitochondrial lineages of Native American origin. Figure 2 shows the map of the central region of Argentina and the relative frequencies of continental lineages in the 20 populations considered, also presented in Table 1. The Native American haplogroups predominate in all surveyed localities, except in Jovita. The greater presence of Eurasian lineages is observed in the plains of the southeast of the region, whereas African lineages are present in 15 of the 20 sampled populations, with higher frequencies in northern Córdoba and the capital city of that province.
Maps showing continental mtDNA haplogroups frequencies in 20 population samples from central Argentina. References for populations abbreviated names are from Table 1
Based on historical records that account for differences in the occupation of the regions of Sierras (mountain ranges) and plains [15], populations were grouped into two groups to assess whether there are differences in the distribution of continental maternal lineages or not. The AMOVA was performed with three variants with respect to the sample of Córdoba city: first, including it in the Sierras group, second including it in the Plains group, and third, excluding it from the analysis. The three results show significant differences between groups (p < 0.001). The maximum differentiation between groups was observed when Córdoba city was not included in the analysis (FST 0.149), although the other configurations also result in high differentiation between groups (FST 0.102 and FST 0.117). The inclusion of Cordoba city decreases this differentiation because it is a large and cosmopolitan city, recipient of many migrants, resulting in an average composition of the two regions.
To further investigate the geographical structure of the genetic variation, we first carried out a Mantel test between matrices of geographical and genetic (FST) distances, calculated based on relative frequencies of continental lineages. A correlation of r2 = 0.463 (p < 0.05) was observed, indicating a significant association. The spatial pattern of continental lineages can be better visualized through the isofrequency maps. Figure 3a shows a clear clinal variation, with higher Native American frequencies in the northwest, decreasing to the southeastern direction; whereas a similar opposite patterns is observed for European lineages with higher frequencies in the southeast and lower frequencies in the northeast and west, particularly in the villages of San Luis and southern Santiago del Estero (Fig. 3b). In Fig. 3c, on the other hand, it is observed that the African lineages are concentrated mainly in the Sierras region of Córdoba and in the capital of that province.
Isoline maps of central Argentina showing spatial frequency distribution of continental mtDNA haplogroups. a Native America lineages. b European lineages. c African lineages. References in Fig. 1
The four major haplogroups (A2, B2, C1, and D1) of Native American origin are present in all the populations studied (Table 1). C1 and D1 are the most frequent haplogroups in the provinces of Santiago del Estero and Córdoba, except Amboy and Cruz Alta, where haplogroup B2 is the most common. In San Luis province, the distribution is more heterogeneous between the four sampled localities, being C1 and A2 the haplogroups most common in the entire province.
Native American mitochondrial lineages in central Argentina
We obtained sequences of the entire mtDNA control region from 334 samples that present haplogroups of Native American origin. In another 181 samples presenting Native American lineages, it was only possible to obtain partial control region mtDNA sequences (Supplementary Table S1). The genetic structure of the population was investigated by AMOVA based on the information of the HVRI mtDNA region between 16,027 and 16,362 bp. The sample of San Carlos Minas, with low sample size, was grouped together with Villa de Soto, geographically close. The first analysis, considering all populations together, shows that 98.13% of genetic variation occurs within populations, whereas only 1.87% reflects the variation among populations. The observed FST is low but still statistically significant (0.018; p = 0.005). Then, we carried comparisons among provinces (Córdoba/San Luis/Santiago del Estero), and between geographical regions (sierras/plains). Both configurations represent low and no significant differences between groups (FST: 0.004, p = 0.118, and FST: 0.001, p = 0.398, respectively).
Diversity parameters were calculated based on mtDNA complete control region (positions from 15,950 to 546 bp). Summary diversity estimators are showed in Table 3. The highest genetic diversity rate (Ĥ) was detected in Córdoba province, but the other two provinces also present high levels of genetic diversity. Similarly, nucleotide diversity levels are high in all three provinces, with an average value for central Argentina of 0.012 ± 0.006. On the other hand, negative and significant values were observed in the neutrality test (Tajima’s D and Fu’s FS) when Central Argentina is consider a single sample set, indicating a relatively recent population growth.
Native American mtDNA variation in the Southern Cone
The AMOVA, performed including 47 populations of the Southern Cone of South America, shows that almost 12% of the genetic variation for the mtDNA HVRI corresponds to the interpopulational component (FST = 0.117, p = 0.000). Differentiation among regions is larger than differentiation among populations within groups (FCT = 0.073 vs. FSC = 0.048), both values being significantly different from 0 (p < 0.001). This result shows that the genetic variation is strongly structured geographically, at least in relation to maternal lineages.
We performed an eigen-analysis based on the distance matrix estimated from pairwise genetic distances K2P. Seven independent vectors accumulates >99% of the total variation. These derived variables were in turn used as input for the DA among regional groups presented in Table 2, and previously analyzed by mean of the AMOVA. DA shows strong geographical structure of the mtDNA variation in the Southern Cone. According to the Mahalanobis distance to the centroid of the group, on average, 82.3% of the population samples were classified into their original group. Samples from central Argentina, subtropical forests, and Patagonia and Tierra del Fuego regions were all correctly classified. Seven of nine populations of the central Andes were also well classified, whereas the remaining two were assigned to the group NWA, also constituted by populations of Andean origin and living at high altitude environments. Three of the seven populations of Cuyo region were classified as belonging to central Argentina, most probably due to the geographic proximity of these two regions and the numerous haplotypes shared [52, 53]. Three of the four populations of the lowland Bolivia were correctly classified and the remaining population was grouped along with NWA. Unsurprisingly, the Gran Chaco, located at the center of the Southern Cone, is the more dispersed group with only three of the five populations correctly. Note that we also performed the classification by means of the Jackknife procedure. Although under this alternative approach, the percentage of correct classification was reduced to 53.2% (still much higher than the 12.5% expected by chance alone), all the misclassified populations still belong to neighbor regions with similar evolutionary histories (i.e., NWA Andes Central Argentina and Cuyo).
This strong geographic pattern is well illustrated in Fig. 4, where the population samples and the centroid groups are projected onto the first two discriminant functions, representing 48.5% and 30.4% of the total variation, respectively. The populations from central Argentina appear very close to each other at the center of the arrangement and are surrounded by the samples from Cuyo, forming together a single cluster that reflects geographical closeness and likely a common origin [52, 53]. Patagonia and Tierra del Fuego populations, by one side, and subtropical forests groups by the other, constitutes two clusters sharply separated from each other and from the other groups. Andean samples are grouped on the left side of the plot and overlap with the NWA populations (also of Andean origin), and to a lesser extent to the Gran Chaco. The lowlands of Bolivia samples, are more scattered across the graph.
Discriminant analysis of principal coordinates vectors extracted from K2P distances. Populations and their abbreviated names are presented in Table 2. Centroids of geographical regions in bold. Central Argentina (CARG): filled circles; western Argentina (CUYO): open circles; Gran Chaco (CHACO): squares; Patagonia and Tierra del Fuego (PTF): diamonds; northwestern Argentina (NWA): triangles; subtropical forest (FOREST): filled squares; Central Andes (ANDES): filled triangles; Lowlands Bolivia (LLB): filled diamonds. (Color figure online)
Spatial analysis
Mantel test between K2P and geodesic distance matrices reveals moderate but significant geographic structure of mtDNA variation (r = 0.274, p = 0.001). The Mantel correlogram shows that this correlation is significantly positive up to 1500 kms, declining to insignificant values after that (Fig. 5a). The bearing analysis (Fig. 5b) reveals two patterns of stronger positive correlations: the first is observed at 5° (roughly E-W, r = 0.251, p = 0.003) and explains gene flow between central Argentina and Cuyo populations. The second is observed at 150° (roughly NW-SE, r = 0.247, p = 0.003) and accounts for gene flow between the Andean region and NWA and Gran Chaco populations.
Analysis of paternal lineages
Continental origin of paternal lineages
Among the 258 male samples analyzed in this work, we observed five Y chromosome haplogroups/paragroups distributed fairly homogeneously across the three provinces (Table 4). R1 is the most common haplogroup in the studied populations and in European populations as previously described [40]. The next most frequent clade is F(xK), widely distributed in Europe, Asia, and North Africa. Twenty-seven individuals display the haplogroup D–E, widely distributed in Africa, Middle East, and Europe. Only three individuals carry the subhaplogoup E1b1a, which is the major Y chromosome haplogroup found in Africa [38]. Another three individuals belong to the para-haplogroup K(xQ,R), widely distributed in Asia, Africa, and Europe [40]. Finally, Q1a3a (Q-M3), the principal Y chromosome haplogroup of American origin, is present in higher proportion in Santiago del Estero (11.1%), followed by San Luis (10.3%), and is poorly represented in Cordoba (5.2%). This haplogroup is defined by the markers M242, M346, and M3. The para-haplogroup clade Q1a3*, presenting the sites M242 and M346, is found in some populations of North America, at low frequencies in Central and South America, and is absent in populations from central Argentina [24, 39]. In summary, the populations studied in this work possess, in average, 92% male lineages of European/ Middle Eastern origin, 1% of African origin, and 7% of Native American origin. In this approach, we might be underestimating the African component, considering lineages E(xE1b1a) as Europeans, whereas other work on Argentine urban populations indicates that the total percentage of male lineages of African origin is very low, around 1.7% [54].
Regional structure
The analysis of genetic structure based on paternal lineages was carried out between provinces and geographic regions due to low samples size at the level of local populations. The AMOVA reveal no genetic differentiation in the distribution of paternal lineages among provinces (Córdoba/San Luis/Santiago del Estero, FST: −0.034, p = 0.978) nor between geographic regions (sierras vs plains, FST: −0.003, p = 0.446). These results show a male population with diverse continental origins but homogeneously distributed throughout the central Argentina region.
Interpopulational analysis of Y chromosome haplogroups
In order to investigate the distribution of paternal lineages of central Argentinian populations in a wider geographical context, we included in the analysis other native and admixed populations from Argentina, Chile, and Paraguay, taking the information provided by the article of Bailliet et al. [24]. A summary of the populations considered is presented in Table 4. Samples from the Gran Chaco, Patagonia, and northwestern Jujuy (pooled from Susques, Cochinoca, and Rinconada, three small towns located above 3500 m of altitude), are of Native American origin, whereas all the other samples were obtained in admixed populations. The multidimensional scaling (MDS) plot of Fig. 6, shows the native population samples, with higher incidence of Q haplogroups, on the left side of axis 1, whereas the admixed populations fall along the right side of same axis, due to high frequencies of allochtonous haplogroups, principally R1 and F. Samples from central Argentina cluster together close to Tucumán, Mendoza, Catamarca, and La Rioja, all urban populations from northern and western Argentina. The samples from Tartagal and Salta, with relatively high values of Q1a3a lineages, fall on intermediate position between native and urban populations, as well as Huilliche, Mapuche, and Mocoví, showing that these native groups have high frequencies of non Native American paternal lineages. The second axis separates Ayoreo and Lengua, two native populations from the Paraguayan Chaco, from the other native samples, due to their high incidence of paragroup Q1a3*. Azampay and Aguaray, with very high frequencies of R1 and low in F haplogroups, split from the other admixed samples to the bottom right quadrant of the plot.
MDS plot of chord distances based on Y chromosome haplogroups frequencies. Stress = 0.032. The abbreviations are presented in Table 3. Central Argentina: filled circles; western Argentina: open circles; Gran Chaco: squares; Patagonia: diamonds; northwestern Argentina: triangles
Discussion
Valuable information about the processes of colonization, dispersal, and subsequent evolution of human populations are gathered from different disciplines such as history, demography, archeology, and genetics. The human colonization of the Americas has been extensively studied and discussed from each of these disciplines and, in many cases, through interdisciplinary approaches. However, diversification and regional or local interaction among populations are often poorly documented. The main objective of this work was to analyze, using molecular markers of uniparental inheritance, the genetic composition of the current human populations of three provinces of central Argentina in order to contribute to the understanding of micro-evolutionary and historical processes that gave rise to the observed genetic diversity.
During the construction of the Argentinean Nation, governments privileged an ideological process that denied the existence of non-European background in the country’s population [55]. This process resulted in projecting the image of a country made up from a “melting pot”, but only in reference to the contribution of different European segments, in the consolidation of the representation of a “national being” as part of a civilized and capitalist world, as opposed to “others”, meaning Native Americans and Africans [56, 57]. According to the official history, Native Americans and Africans disappeared due to wars, epidemics, and miscegenation. However, studies including historical demography [58] and anthropological genetics [16, 22, 54, 59, 60], show that this “disappearance” rather than indicating a change in the genetic composition of the population represents the efforts of the government politics for a process of cultural “whitening” the society. The mechanism employed to replace the native population with European immigrants did not occur with the same intensity throughout Argentina. The immigration waves had less impact in the north–central and northwestern regions, the most populated areas of the country in pre-Hispanic times. The spatial structure of genetic diversity has its origins in historical factors. It is possible to distinguish different stages in migratory processes from abroad, with a heterogeneous regional impact. The genetic composition of central Argentina gives account of these processes. On one hand, the political boundaries between provinces influenced the configuration of the genetic structure of the populations that were formed. In this sense, Córdoba—an important economic and commercial center since colonial times—has a greater component of foreign lineages than the populations of San Luis and Santiago del Estero. On the other hand, the genetic structure of central Argentina also accounts for other processes related to different migration phases and occupations of space over the last 500 years. In colonial times, the population was concentrated in Córdoba city and along the mountain ranges. The Sierras region, which concentrated most of the native population settlements before the arrival of the Spaniards, was also the place where we have found the highest proportions of Native American and African maternal lineages. In contrast, the region of the plains (Pampas) represents the most sparsely native population occupation and, together with the large cities, the principal destination of the European migrants during 1880–1930 period. This is reflected in the higher proportion of maternal Eurasian lineages in populations of southern Córdoba, as well as in other towns of the Pampas region [54, 61, 62]. Our results contrast with those obtained by Catelli et al. [62], and Salas et al. [63], who observed a much lower incidence of Native American maternal lineages in samples gathered from Córdoba province (41% and 47%, respectively). Those lower prevalences most likely reflect differences in the sample colletion. Both authors used samples of urban origin collected in different forensic, anthropological, or clinical laboratories, using different sampling criteria.
Although Córdoba city has traditionally been regarded as one of the large “white” city of Argentina [64], our results show high incidence of maternal native lineages in the north and western areas of that province, almost at the level found in Santiago del Estero and San Luis, two provinces that have received less European immigration and show extremely high prevalence of Native American maternal lineages (92.9% and 87.2% respectively).
This differentiation between geographical regions is clear in relation to the distribution of continental mitochondrial lineages, but not regarding patrilineages, which show homogeneous distribution across the landscape. Thus, populations of central Argentina show the same Y chromosome haplogroup distribution than other populations such as Catamarca, La Rioja, Mendoza, and Tucumán, indicating analogous migration processes, with a distribution similar to western European populations, and with a very low incidence of Native American lineages [24]. One of the most obvious patterns observed in this study is the discrepancy between the maternal and paternal lineages, involving a sex-asymmetric gene flow primarily involving European men and African and American women [20, 21]. These results, recurrent in contemporary Latin American populations, are also observed in populations from central Argentina in concordance with the pattern observed by Catelli et al. [62], and Salas et al. [63] in samples of Córdoba province. The exception to this pattern is the population of Jovita, where most of the maternal and paternal lineages are of European origin.
It is worth noting the high incidence of African mitochondrial lineages in some villages of the Sierras of Cordoba with respect to other Argentine populations. This region was one of the major settlement locations of African people during the colonial period, associated with Jesuit missions and Jesuit productive establishments [65].
The AMOVA results performed at regional level based on the mtDNA HVR-I show that both configurations considered (between provinces and between plain and mountain regions) do not account for significant differentiations between groups. Similarly, the distribution pattern of the four Amerindian mitochondrial haplogroups is very similar in the three provinces of central Argentina. The high and similar levels of genetic diversity found in Córdoba, Santiago del Estero, and San Luis provinces, on the other hand, would indicate that they were subjected to similar evolutionary forces capable of maintaining a high genetic variability in the total population. From the evolutive point of view, for neutral loci such as mitochondrial DNA polymorphisms, maintaining genetic variability depends directly on the effective size of the population [66]. The existence of sporadic events of gene flow from communities between the populations of central Argentinian or from neighboring regions, in pre-Columbian times and, mainly, after the arrival of the Spaniards, would have contributed to the increase of the effective size of the population [67].
With the aim of analyzing the genetic variation of the populations of central Argentina on a larger geographic scale, we carried out analyzes taking into account other populations of the Southern Cone of South America. The populations of central Argentina show genetic [52, 53], and morphological [68, 69] similarities with the populations from Cuyo (western Argentina) and other populations of the Sierras Pampeanas (Catamarca and La Rioja), with distances not statistically significant. This genetic affinity does not agree with the traditional model of settlement, according to which the origin of the native populations of central Argentina, both from archaeological and bioanthropological evidence, would be closely linked to the central Andean region [5, 70, 71], a model supported by other authors in recent works [72].
In spite of the genetic affinities with other populations of the Southern Cone, it is also possible to observe particular and exclusive genetic characteristics in central Argentina, which differentiates it from the rest of the populations. A possible hypothesis to explain the homogeneity in the distribution of genetic variability is that this pattern would be the result of differential permeability between local groups and those coming from other geographic regions of Argentina. Thus, the genetic diversity with high homogeneity observed throughout central Argentina for maternal lineages might be reflecting a fluid gene flow between local populations and resistance for receiving new contributions from groups from other regions. In the particular case of the province of Cordoba, archaeological studies report the existence of links and cooperation among different peoples who occupied that region in pre-Hispanic times [73, 74]. Those relationships would have allowed exchanges that conducted to the genetic homogeneity in the territory of that province. According to earlier studies of our group based on mtDNA sequences and autosomal AIMS, we observed in this study a large genetic homogeneity among populations in central Argentina [17, 22, 53]. However, we also observed that haplogroup B2 segregates at higher frequency in three populations (Amboy, Cruz Alta, and Tilisarao). In this sense, it is worth to mention that a high relative prevalence of haplogroups C1 and D1 occurs in ancient samples (1192 ± 40 to 370 ± 15 14C years before present) of the plains of Córdoba province, whereas B2 is the most frequent in Sierras ancient samples (1080 ± 40 to 345 ± 20 14C years before present) [6, 8], which might be reflecting continuity with past inhabitants in some cases, population replacements in others, and/or recent migratory events.
Similarly, negative values observed in the neutrality tests (Tajima’s D and Fu’s FS), indicate relatively recent population growth, probably associated with technological and organizational changes leading to new lifestyles and important demographic and territorial expansion [75]. In conclusion, the molecular markers of maternal inheritance shows large genetic diversity homogeneously distributed throughout central Argentina, probably as result of a recent common origin and sustained gene flow between sub-populations. In addition, migratory events that occurred in colonial and recent times should have contributed to hiding any traces of differentiation that might have existed in the past. The analysis of paternal lineages showed also homogeneous distribution of the variation across the region but also a drastic reduction of the native male population, with a large prevalence of haplogroups of European origin.
Finally, our results highlight the need to rethink common-sense assumptions, which generally refer to the absence or complete extinction of Native Americans and Africans in Argentina, with a detachment of the official history from its pre-Hispanic past. And yet, this past is still present in the people’s genomes.
References
Rivero D, Roldán F. Initial peopling of the Córdoba mountains, Argentina: first evidence from El Alto 3. Curr Res Pleistocene. 2005;22:33–35.
Laguens AG, Pautassi EA, Sario GM, Cattáneo GR. ELS1, a fishtail projectile-point site from central Argentina. Curr Res Pleistocene. 2007;24:55–57.
Cornero S, Neves WA, Prous A. Prevalencia de caries en una muestra de Santana Do Riacho o el mito del paleoindio cazador. Rev Argent Antropol Biológica. 1999;2:301–6.
González RA, Pérez Gollán JA. Argentina indígena, vísperas de la conquista. (Editorial Paidós, Buenos Aires, Argentina, 1976).
Serrano A. Los Comechingones. Serie Aborígenes Argentinos. Instituto de Arqueología, Lingüística y Folklore, Universidad Nacional de Córdoba I, (1945).
Nores R, Fabra M, Demarchi DA. Variación temporal y espacial en poblaciones prehispánicas de Córdoba. Análisis de ADN antiguo. Rev Del Mus Antropol. 2011;4:187–94.
Fabra M, Demarchi DA. Análisis morfogeométrico aplicado al estudio de los patrones espaciales y temporales de variación morfológica craneofacial en poblaciones del centro de Argentina. In: Revista Cuadernos del Instituto Nacional de Pensamiento Latinoamericano, Series Especiales.1° Congreso Internacional de Arqueología de la Cuenca del Plata 1:87-101. 1, (2013).
Rodrigo N, Fabra M, García A, Demarchi DA. Diversidad genética en restos humanos arqueológicos del sitio El Diquecito (costa sur, Laguna Mar Chiquita, provincia de Córdoba). Rev Argent Antropol Biológica. 2017;19:1–12.
Laguens A, Fabra M, Dos Santos GM, Demarchi DA. Paleodietary inferences based on isotopic evidences for populations of the Central Mountains of Argentina during the Holocene. Int J Osteoarchaeol. 2009;19:237–49.
Fabra M, Nores R, Salega S, González C. In: Avances recientes en la Bioarqueología Sudamericana (ed. Luna, L Aranda, CYJS) 205-31 (Ciudad Autónoma de Buenos Aires: Grupo de Investigación de Bioarqueología, 2014).
Fabra M, González C, Robin S. Evidencias de violencia interpersonal en poblaciones del piedemonte y las llanuras de Córdoba (Argentina) a finales del Holoceno tardío. RUNA. 2015;36:5–27.
Fabra M, González CV. Diet and oral health of populations that inhabited central Argentina (Córdoba province) during Late Holocene. Int J Osteoarchaeol. 2015;25:160–75.
Palomeque S. In: Nueva historia Argentina, Tomo 2. (ed. Tandeter, E) 87–144 (Editorial Sudamericana, Buenos Aires, Argentina, 2000).
Pellegrino A. La migración internacional en América Latina. Tendencias y perfiles de los migrantes. Santiago de Chile (2002).
Tell S. Córdoba rural, una sociedad campesina (1750-850). (2008).
García A, Demarchi DA. Incidence and distribution of Native American mtDNA haplogroups in central Argentina. Hum Biol Int Rec Res. 2009;81:59–69.
García A. Historia evolutiva de las poblaciones originarias del actual territorio de la provincia de Córdoba: Evidencias moleculares. (Universidad Nacional de Córdoba, 2011).
Pauro M, García A, Nores R, Demarchi DA. Analysis of uniparental lineages in two villages of Santiago Del Estero, Argentina, seat of Pueblos de Indios in colonial times. Hum Biol. 2013;85:699–720.
García A, Demarchi DA. Linajes parentales amerindios en poblaciones del norte de Córdoba. Rev Argent Antropol Biológica. 2006;8:57–71.
Mesa NR, et al. Autosomal, mtDNA, and Y-chromosome diversity in Amerinds: pre- and post-Columbian patterns of gene flow in South America. Am J Hum Genet. 2000;67:1277–86.
Wang S, et al. Geographic patterns of genome admixture in Latin American Mestizos. PLos Genet. 2008;4:e1000037 https://doi.org/10.1371/journal.pgen.1000037.
García A, et al. High interpopulation homogeneity in Central Argentina as assessed by ancestry informative markers (AIMs). Genet Mol Biol. 2015;38:324–31.
García A, Pauro M, Nores R, Bravi CM, Demarchi DA. Phylogeography of mitochondrial haplogroup D1: an early spread of subhaplogroup D1j from Central Argentina. Am J Phys Anthropol. 2012;149:583–90.
Bailliet G, et al. Antecedentes y nuevos aportes en el estudio del Cromosoma Y en poblaciones humanas sudamericanas. J Basic Appl Genet. 2011;22:1–9.
Stone AC, Stoneking M. Ancient DNA from a pre-Columbian Amerindian population. Am J Phys Anthropol. 1993;92:463–71.
Umetsu K, Yuasa I. Recent progress in mitochondrial DNA analysis. Leg Med 7. 2005;7:259–62.
Andrews RM, et al. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147.
Weissensteiner H, et al. HaploGrep 2: mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucl Acids Res. 2016. https://doi.org/10.1093/nar/gkw233.
van Oven M. PhyloTree Build 17: growing the human mitochondrial DNA tree. Forensic Sci Int Genet Suppl Ser. 2015;5:9–11.
Long F, Yong-Gang Y. An update to MitoTool: using a new scoring system for faster mtDNA haplogroup determination. Mitochondrion . 2013;13:360–3.
Underhill PA, Jin L, Zemans R, Oefner PJ, Sforza LLC. A pre-Columbian Y chromosome-specific transition and its implications for human evolutionary history. Proc Natl Acad Sci USA. 1996;93:196–200.
Su B, et al. Y-chromosome evidence for a Northward migration of modern humans into Eastern Asia during the last ice age. Am J Hum Genet. 1999;65:1718–24.
Underhill PA, et al. Maori origins, Y-chromosome haplotypes and implications for human history in the Pacific. Hum Mutat. 2001;17:271–80.
Karafet TM, et al. Ancestral Asian source (s) of new world Y-chromosome founder haplotypes. Am J Hum Genet. 1999;64:817–31.
Seielstad M, et al. Construction of human Y-chromosomal haplotypes using a new polymorphic A to G transition. Hum Mol Genet. 1994;3:2159–61.
Hammer MF. A recent insertion of an Alu element on the Y chromosome is a useful marker for human population studies. Mol Biol Evol. 1994;11:749–61.
Jurado Medina LS, Muzzio M, Bravi Constantino ML, Barreto G, Bailliet G. Short communication human Y-chromosome SNP characterization by multiplex amplified product-length polymorphism analysis. Electrophoresis. 2014;35:2524–7.
Hammer MF, et al. Hierarchical patterns of global human Y-chromosome diversity. Mol Biol Evol. 2001;7:1189–203.
Karafet TM, et al. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 2008;18:830–8.
Jobling MA, Tyler-smith C. The human Y chromosome: an evolutionary marker comes of age. Nat Rev Genet. 2003;4:598–612.
Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application. Genetics. 1992;491:479–91.
Excoffier L, Lischer H. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–7.
Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–20.
Gower JC. Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika. 1996;53:325–38.
Jombart T, Devillard S, Balloux F. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 2010;11:94.
Mantel N. The detection approach of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–20.
Smouse PE, Peakall ROD. Spatial autocorrelation analysis of individual multiallele and multilocus genetic structure. Hered (Edinb). 1999;82:561–73.
Falsetti AB, Sokal RR. Genetic structure of human populations in the British Isles. Ann Hum Biol. 1993;20:215–29.
Rosenberg MS, Anderson CD. PASSaGE: pattern analysis, spatial statistics and geographic Exegesis. Version 2. Methods Ecol Evol. 2011;2:229–32.
Kruskal JB. Multidimensional scaling by optimizing goodness of fit to a nonmetric hypothesis. Psychometrika. 1964;29:1–27.
Hammer Ø, Harper DAT, Ryan PD. PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron. 2001;4:1–9.
Motti JMB. Caracterización de linajes maternos en la población actual del noroeste y centro-oeste argentinos. (Universidad Nacional de La Plata, 2012).
Pauro M. Análisis molecular de linajes uniparentales en poblaciones humanas del centro de Argentina. (Universidad Nacional de Córdoba, 2015).
Corach D, et al. Inferring continental ancestry of argentineans from autosomal, Y-chromosomal and mitochondrial DNA. Ann Hum Genet. 2010;74:65–76.
Chamosa O. Indigenous or Criollo: the myth of white Argentina in Tucumán’s Calchaquí valley. Hisp Am Hist Rev. 2008;88:71–106.
Bonfil Batalla G. Participación en la Mesa Marxismo y Antropología. In: Nueva Antropología año III, 3–59 (1979).
Lins Ribeiro G. In: La Antropología brasileña contemporánea. Contribuciones para un diálogo latinoamericano (eds. Grimson, Lins Ribeiro & Semán) 165-95 (Prometeo, Buenos Aires, Argentina, 2004).
Andrews GR. The afro-argentines of Buenos Aires, 1800-1900 (The University of Wisconsin press, London, England, 1980).
Avena S, et al. Mestizaje en el sur de la Region Pampeana (Argentina). Su estimación mediante el análisis de marcadores proteicos y moleculares uniparentales. Rev Argent Antropol Biol. 2007;9:59–76.
Pauro M, Bravi CM. Distribución de haplogrupos mitocondriales alóctonos en poblaciones rurales de Córdoba y San Luis. Rev Argent Antropol Biol. 2010;12:47–55.
Bobillo MC, et al. Amerindian mitochondrial DNA haplogroups predominate in the population of Argentina: towards a first nationwide forensic mitochondrial DNA sequence database. Int J Leg Med. 2010;124:263–8.
Catelli ML, et al. The impact of modern migrations on present-day multi-ethnic Argentina as recorded on the mitochondrial DNA genome. BMC Genet. 2011;12:77.
Salas A, Jaime JC, Alvarez-Iglesias V, Carracedo A. Gender bias in the multi-ethnic genetic composition of central Argentina. J Hum Genet. 2008;53:662–74.
Grosso JL. Indios Muerto, Negros Invisibles. (Encuentro Grupo Editor. Serie Inter/Cultura. Memoria y Patrimonio. Córdoba, Argentina, 2008).
Becerra MJ, Buffa D, García C, Vagni JJ, Zeballos JM. Pasado y Presente del aporte africano en la identidad cordobesa contemporánea. In: III Congreso de la Asociación Latinoamericana de Población, ALAP 1–13 (2008).
Kimura M. Evolutionary rate at the molecular level. Nature. 1968;217:624–6.
Gaya-Vidal M, et al. mtDNA and Y-chromosome diversity in Aymaras and Quechuas from Bolivia: different stories and special genetic Ttraits of the Andean Altiplano populations. Am J Phys Anthropol. 2011;142:215–30.
Fabra M. Relaciones biológicas craneales entre poblaciones prehispánicas de la región central del territorio argentino y otras del Cono Sur sudamericano. Rev An Arqueol Y Etnol la Fac Filos Y Let la Univ Nac Cuyo. 2008;61-62:123–45.
Fabra M, Demarchi DA. Variabilidad craneofacial en poblaciones del sector austral de las Sierras Pampeanas: aportes desde la morfometría geométrica. Rev Relac la Soc Argent Antropol. 2009;34:1–34.
Outes F. Los tiempos prehistóricos y protohistóricos en la Provincia de Córdoba. Sep la Rev Del Mus la Plata. 1911;XVII:261–374.
González AR. Algunas observaciones sobre los caracteres antropológicos de los primitivos habitantes de Córdoba. Publ Del Inst Arqueol Lingüíst Y Folk Univ Nac Córdoba. 1944;31:1185–90.
Rivero DE, Berberián EE. El poblamiento inicial de las Sierras Centrales de Argentina. Las evidencias arqueológicas tempranas. Cazadores Recolectores del Cono Sur. Rev Arqueol. 2006;1:127–38.
Piana J. Los indígenas de Córdoba bajo el régimen colonial 1570-620. Ethnohistory. 1995;42:193–5.
Pastor S, Medina M, Recalde A, Berberián EE. Arqueología de la región montañosa central de argentina. Avances en el conocimiento de la historia prehispánica tardía. Relac Soc Argent Antropol. 2012;37:89–112.
Laguens AG, Bonnín M. Sociedades indígenas de las Sierras Centrales: arqueología de Córdoba y San Luis (Universidad Nacional de Córdoba, Córdoba, Argentina, 2009).
Acknowledgements
We especially thank the participants for their cooperation with our work during the collection of biological samples. Thanks are due to the Mariana Fabra and Pierre Luisi for useful comments on the manuscript. Angelina García, Graciela Bailliet, Claudio Bravi, and Darío Demarchi are Research Career Member of the Consejo Nacional de Investigaciones Científicas y Técnicas de la República Argentina (CONICET). This investigation was funded by ANPCyT through grants PICT 2012-711 and by SECyT (Universidad Nacional de Córdoba).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
10038_2017_406_MOESM1_ESM.xlsx
Table S1. Polymorphic sites in mtDNA complete Control Region defining Native American haplogroups detected in three Central Argentinanian populations. The absolute frequency per haplotype in each population
Rights and permissions
About this article
Cite this article
García, A., Pauro, M., Bailliet, G. et al. Genetic variation in populations from central Argentina based on mitochondrial and Y chromosome DNA evidence. J Hum Genet 63, 493–507 (2018). https://doi.org/10.1038/s10038-017-0406-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-017-0406-7
This article is cited by
-
Ancestral genetic legacy of the extant population of Argentina as predicted by autosomal and X-chromosomal DIPs
Molecular Genetics and Genomics (2021)
-
Sex-biased patterns shaped the genetic history of Roma
Scientific Reports (2020)
-
Demand for larger Y-STR reference databases in ethnic melting-pot countries: Argentina as a test case
International Journal of Legal Medicine (2019)