Abstract
Although the aetiology of non-syndromic cleft lip with or without cleft palate (nsCL/P) has been studied extensively, knowledge regarding the role of genetic factors in the pathogenesis of this common craniofacial anomaly is still limited. We conducted a follow-up association study to confirm that CDKAL1 nucleotide variants identified in our genome-wide association study (GWAS) for nsCL/P are associated with the risk of this anomaly. In addition, we performed a sequence analysis of the selected CDKAL1 exons. A mega-analysis of the pooled individual data from the GWAS and a replication study revealed that six out of thirteen CDKAL1 variants were positively replicated and reached the threshold of statistical significance (Ptrend < 3.85E−03). They represented a single association signal and were located within the fifth intron of CDKAL1. The strongest individual variant was rs9356746 with a Ptrend value = 5.71E−06 (odds ratio (OR) = 1.60, 95% confidence interval (CI): 1.30–1.97). Sequencing analysis did not reveal any pathogenic mutations of this gene. This study provides the first evidence that chromosomal region 6p22.3 is a novel susceptibility locus for nsCL/P. The location of the risk variants within the CDKAL1 intronic sequence containing enhancer elements predicted to regulate the SOX4 transcription may suggest that SOX4, rather than CDKAL1, is a potential candidate gene for this craniofacial anomaly.
Similar content being viewed by others
Introduction
Genome-wide association studies (GWASs) are a powerful tool for investigating the role of genetic factors in the aetiology of common human diseases. Over the last 12 years they have identified a large number of loci associated with disease outcomes and have provided important insights into the pathogenesis of various diseases and conditions [1, 2]. It is worth noting that the vast majority of disease risk variants detected by GWASs are located within non-coding regions of the genome and their biological function is largely unknown [3]. Within the field of birth defects, recent GWASs have significantly increased the knowledge about the genetic architecture of non-syndromic cleft lip with or without cleft palate (nsCL/P, OMIM %119530), which is the most common craniofacial anomaly, with an overall birth prevalence of 1 per 700 live births [4]. To date, seven independent GWASs for nsCL/P have been conducted. These have identified several novel cleft-susceptibility loci and novel candidate genes, including 1p22.1 (ARHGAP29), 2p24.2 (FAM49A), 8q24.21 (gene desert), 10q25.3 (VAX1), 12q12 (ADAMTS20), 16p13.3 (ADCY9), 17q22 (NOG), 17q23 (TANC2), 19q13 (RHPN2) and 20q12 (MAFB) [5,6,7,8,9,10,11]. In these studies the most consistent results were observed for nucleotide variants in the gene-poor region of chromosome 8q24.21. Studies in mice have shown that this locus contains very distant cis-acting enhancers that control Myc expression in the developing face [12].
A GWAS for nsCL/P was also conducted in a homogenous Polish population (unpublished results). In this study we found that nucleotide variants located within the large fifth intron of the CDKAL1 gene (CDK5 regulatory subunit-associated protein 1 Like 1, OMIM *611259) are associated with an increased risk of this craniofacial anomaly. These results were not statistically significant. However, they were close to the suggestive genome-wide significance level (Ptrend < 1.00E−05).
CDKAL1 is one of the major candidate genes reproducibly associated with type 2 diabetes mellitus (T2DM) [13,14,15,16]. Interestingly, single-nucleotide polymorphism (SNPs) associated with T2DM in European and Asian populations have also been mapped to intron 5 of CDKAL1 [13]. It has been demonstrated that these diabetes risk variants are located within the linkage disequilibrium (LD) block containing highly conserved non-coding elements that are likely to regulate SOX4 transcription [17]. Since SOX4 (SRY-box 4, OMIM *184430) is a regulatory gene that has already been proposed as a candidate gene for orofacial clefts [18], we decided to conduct a follow-up association study to confirm that the CDKAL1 variants identified in our GWAS are associated with the risk of nsCL/P. In addition, we performed a sequence analysis of the selected CDKAL1 exons, in order to detect rare risk variants potentially implicated in the aetiology of this structural anomaly.
Materials and methods
Study design
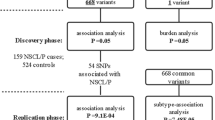
The study was composed of four stages: (I) a statistical analysis of common SNPs (n = 245) located within the CDKAL1 gene and adjacent regions genotyped in our case–control GWAS for nsCL/P, (II) selection and genotyping of the CDKAL1 top-ranked SNPs (n = 13) in the independent group of nsCL/P patients and controls, (III) a statistical analysis using data from the replication cohort and a combined analysis using pooled data from GWAS and replication cohorts, and (IV) mutation screening of CDKAL1 exons 3 to 7 in patients with nsCL/P.
Study population
All the study participants were unrelated Caucasians of Polish origin. The study protocols were approved by the Institutional Review Board of Poznan University of Medical Sciences [19]. Informed consent was obtained from all individuals enroled in the study, or their legal guardians. The patients with a diagnosis of nsCL/P were recruited from several Polish medical centres. Case eligibility was ascertained by clinicians using detailed diagnostic information from the medical records. The control group was composed of healthy individuals without any developmental anomalies and with no family history of congenital disorders. After stringent quality control (QC) the GWAS cohort consisted of 269 nsCL/P patients (58.0% males) and 569 controls (49.6% males). The replication cohort included 240 nsCL/P patients (57.9% males) and 445 controls (49.9% males). Mutation screening was conducted on 55 patients with nsCL/P (56.4% males). In the patient group, the percentage of individuals with non-syndromic cleft lip only (nsCLO) was 19.6%. Detailed characteristics of all the study participants are presented in the Supplementary Table 1. Genomic DNA was isolated from peripheral blood lymphocytes with the salting-out method.
Replication SNP selection and genotyping
The genotyping results for common SNPs (minor allele frequency, MAF ≥ 0.05) located within the CDKAL1 gene and adjacent regions (±100 kb) were retrieved from our GWAS data (Supplementary Table 2). Genome-wide genotyping was performed using the HumanOmni ExpressExome-8 v1 array (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. All these 245 SNPs passed stringent QC criteria, including a SNP call rate > 0.95, Hardy–Weinberg equilibrium P value > 0.001 in the controls and the visual inspection of the cluster plots. Selection of the CDKAL1 SNPs for the replication analysis was based on the GWAS association results (Cochran–Armitage trend test), the LD patterns observed and the structure of haplotype blocks across the CDKAL1 gene (Supplementary Table 3). The characteristics and location of the assayed nucleotide variants (n = 13) are presented in the Supplementary Table 4. Genotyping of SNPs in the replication cohort was carried out by high-resolution melting curve analysis (HRM) on the LightCycler 96 system (Roche Diagnostics, Mannheim, Germany) with the use of 5× HOT FIREPol EvaGreen HRM Mix (Solis BioDyne, Tartu, Estonia). For all SNPs, the genotyping quality was tested by repeat analysis of ~10% of randomly selected samples. The primer sequences and HRM conditions are presented in the Supplementary Table 5.
Statistical analysis
All the calculations were performed using the PLINK software package version 1.06 [20]. The association of the CDKAL1 SNPs with nsCL/P in the GWAS and replication cohorts was tested with the Cochran–Armitage trend test. The odds ratio (OR) and corresponding 95% confidence intervals (95% CIs) were used to assess the strength of the association. P values below 5.00E−08 were considered as genome-wide significant. For the replication purposes, P values below 3.85E−03 (0.05/13 SNPs) were interpreted as being statistically significant. Replication was considered as positive when the Ptrend value of the mega-analysis was smaller than the Ptrend value of the GWA analysis [21]. Additional statistical tests were performed on the pooled individual data from the GWAS and replication cohorts. The independence of the SNP association signals was tested by conditional analysis, where the allelic dosage for a given SNP was added as a covariate in a binary logistic regression model (additive model). Associations of the CDKAL1 SNPs with nsCL/P in male and female groups separately and the effects of the genotype × sex interactions were assessed by logistic regression approach. Haplotype-based association analysis of the CDKAL1 gene, using a sliding window approach, was conducted employing logistic regression. Haplotypes with a frequency below 0.01 were excluded. The global P values were obtained by Omnibus tests jointly estimating all haplotype effects at a location. Statistical significance was assessed using the 10,000-fold permutation test. To evaluate whether the association between CDKAL1 variants and the risk of nsCL/P is cleft-type-dependent, separate analyses were conducted for individuals with nsCLO and non-syndromic cleft lip and palate (nsCLP). To assess whether the calculated cleft-type-specific ORs were significantly different, the frequencies of the tested SNPs were compared between the case subgroups using Armitage’s trend test.
Mutation screening
For 55 patients with nsCL/P, mutation screening of the CDKAL1 (ENST00000274695.8) exons 3–7 and their exon–intron boundaries was performed by direct sequencing. Exon selection was based on the SNP association results and the structure of haplotype blocks across the CDKAL1 gene. Cycle sequencing was performed, according to the manufacturer’s instructions, using a BigDyeTM Terminator v3.1 Reaction Cycle Sequencing Kit and an ABI Prism 3730 capillary sequencer (Thermo Fisher Scientific, IL, USA). For all identified CDKAL1 variants, the allele frequencies in the general population were checked against the 1000 Genomes Project database (EUR population; http://www.internationalgenome.org/) and the Exome Aggregation Consortium (ExAC) database (non-Finnish European population; http://exac.broadinstitute.org/). The putative functional consequences of the identified missense variant were analysed using in silico prediction programmes PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/) and SIFT (http://sift.jcvi.org/). The primer sequences and conditions used for the amplification and sequencing of the CDKAL1 exons are presented in the Supplementary Table 5.
Results
GWA analysis
Twenty-nine common CDKAL1 SNPs genotyped on the SNP array were nominally associated (Ptrend < 0.05) with the risk of nsCL/P (Supplementary Table 2 and Fig. 1). The most significant SNPs in the GWAS data set were located within the fifth intron of the CDKAL1 gene (transcript: ENST00000274695.8). The strongest individual SNP was rs9356746 with a Ptrend value = 5.87E−05 (Table 1). The rs9356746 C allele was associated with a 1.76-fold increased risk of nsCL/P (95%CI: 1.33–2.33). In addition, 12 other CDKAL1 nucleotide variants showed ORs in the same direction and Ptrend values below 1.00E−03.
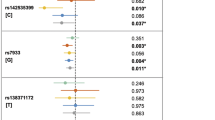
Regional plot of association results within the CDKAL1 locus. The left-hand y-axis shows the Cochran–Armitage trend test P values (−log10 scale) of individual SNPs plotted against their chromosomal position (in Mb) on the x-axis. The right-hand y-axis shows the recombination rate estimated from the HapMap CEU population. The results of both the GWAS (dots) and mega-analysis (squares) are presented. The top SNP in the region, rs9356746, is presented in purple. All other SNPs are colour-coded according to the strength of the pairwise linkage disequilibrium (LD, r2) with the top SNP. The genes in the region, their exon–intron structure, the direction of transcription and the genomic coordinates (according to hg19) are shown at the bottom. Regional plots were generated using the LocusZoom tool version 1.1 [44] (color figure online)
Replication analysis
The top-ranked SNP in the replication data set was rs9356746, with a Ptrend value = 2.03E−02 (Table 1). The OR for the rs9356746 risk allele was 1.43 (95%CI: 1.05–1.94). Two other SNPs (rs9465871 and rs7741604) showed replication Ptrend values < 1.00E−01. None of these results was statistically significant after applying the correction for multiple testing.
Mega-analysis
A mega-analysis of the pooled individual data from the GWAS and replication study confirmed that common CDKAL1 variants are associated with an increased risk of nsCL/P (Table 1 and Fig. 1). Six out of thirteen tested SNPs were positively replicated and showed Ptrend values of a mega-analysis smaller than the Ptrend values found by GWA analysis. All these results remained statistically significant after adjustment for multiple comparisons (Ptrend < 3.85E−03). The allelic ORs for positively replicated SNPs were in the range of 1.26–1.60. For all of them, the minor allele was the risk allele. As in GWAS and replication analysis, the most significant SNP was rs9356746 with a Ptrend value = 5.71E−06 (OR = 1.60, 95% CI: 1.30–1.97). Three other SNPs (rs9465871, rs9358357 and rs7741604) showed Ptrend values < 1.00E−04. These variants were in moderate LD with rs9356746 (r2 values equal to 0.71, 0.68 and 0.56, respectively; Supplementary Table 3). All significant SNPs are located within the fifth intron of CDKAL1 and represent a single nsCL/P association signal (Fig. 1). Their association effects were diminished or abolished when conditioned on each other (Table 2). No significant sex × genotype interactions were observed for nsCL/P (Table 3).
Haplotype analysis
Haplotype analysis revealed several common 2-, 3- and 4-marker CDKAL1 haplotypes associated with nsCL/P (Table 4). These results remained highly significant, even after permutation-based correction. The best evidence of the global haplotype association was detected for haplotypes comprising alleles of rs9358357 and rs9356746 (P = 1.54E−05, Pcorrected = 2.00E−04). The G-C haplotype, consisting of the minor alleles of these nucleotide variants, was associated with a 1.62-fold increase in the risk of nsCL/P compared with the most common haplotype A-T (OR = 1.62, 95% CI: 1.33–2.00, P = 1.95E−06).
Subphenotype analysis
Separate statistical analyses conducted in patients with nsCLP and nsCLO did not reveal any cleft-specific CDKAL1 variants (Table 5). Differences in ORs between the cleft subphenotypes were not statistically significant (heterogeneity P values > 0.05). The most significant SNP identified in this study, rs9356746, was associated with an increased risk of both nsCLP and nsCLO (OR = 1.58, 95% CI: 1.27–1.97 and OR = 1.71, 95% CI: 1.15–2.53, respectively).
Mutation analysis
Sequencing analysis of the CDKAL1 exons 3–7 and their exon–intron boundaries revealed that one patient with nsCLP was a heterozygous carrier of the missense variant, c.116G>A (rs111739077), replacing arginine at position 39 by glutamine (p.Arg39Gln). This rare SNP, predicted to be either deleterious (SIFT) or benign (Poly-Phen2), was one of the nine CDKAL1 missense SNPs tested with the use of the SNP array platform. In the GWAS cohort the rs111739077 variant was identified in three patients with nsCL/P and seven healthy individuals. According to the 1000 Genomes Project and ExAC databases the rs111739077 allele frequency is 0.006 and 0.007, respectively. The other CDKAL1 missense variants tested in the GWAS were not detected in either subjects or controls. Besides the rs111739077 variant, the sequencing analysis identified six common intronic SNPs that were present in either the heterozygous or homozygous state in patients with nsCL/P. Their allele frequencies were similar to those reported in the 1000 Genomes Project (Table 6).
Discussion
The aetiology of nsCL/P is complex and multifactorial, with both genetic and environmental factors contributing to disease risk [4, 22]. Although studied extensively (including GWASs), current knowledge is still not sufficient to explain the molecular pathogenesis of this common developmental anomaly. The present study contributes to a better understanding of the genetic causal factors associated with nsCL/P, since it provides the first evidence that the chromosomal region 6p22.3 might be a novel risk locus for orofacial clefts. We have found that common nucleotide variants of the CDKAL1 gene are significantly correlated with an increased risk of this craniofacial anomaly. In the mega-analysis of pooled data from our GWAS and replication analysis, the top-associated SNP (rs9356746) reached the threshold of suggestive genome-wide significance. There was no evidence of a gender- and cleft-type-dependent association with any of the SNPs studied. A causative role of the CDKAL1 SNPs in the aetiology of nsCL/P was further confirmed by the haplotype analysis, which showed that common haplotypes of this gene may significantly contribute to the risk of this birth defect. All the SNPs tested in this study that fulfilled the criteria of positive replication represent a single association signal and are located within the large fifth intron of the CDKAL1 gene. This pattern of association is similar to that observed for CDKAL1 variants associated with T2DM [13,14,15,16]. The strongest associations with diabetes risk were observed for rs7754840, rs10946398 and rs7756992. However, there is no general consensus to date about any single causal variant [16, 23]. The above-mentioned CDKAL1 variants were also significantly associated with the risk of nsCL/P in our GWA study (P values equal to 2.78E−03, 2.78E−03 and 1.50E−04, respectively).
On the basis of a study by Ragvin et al. [17], the target gene underlying nsCL/P susceptibility at the 6p22.3 locus might be SOX4 rather than CDKAL1, which is probably only a bystander gene. Using comparative genomic analysis they found that CDKAL1 is located within the genomic regulatory block of SOX4 and, along with this downstream gene, is in conserved synteny across vertebrate genomes [17]. In addition, they demonstrated that within the 200-kb LD block, comprising the proximal promoter and exons and introns 1–5 of CDKAL1, the highly conserved enhancer sequences required for the regulation of the SOX4 expression levels are located [17]. This latter gene encodes a TGFβ-regulated transcription factor that performs important functions in developmental processes, including skeletogenesis, embryonic cardiac, thymocyte and nervous system development [24,25,26,27]. Sox4, together with other Sox family members, is critical during neural crest specification, migration and differentiation [28]. In addition, the Sox4 expression profile in the developing mouse palate suggests that it plays a number of functional roles during palatogenesis. These include contributions to medial edge epithelium fusion and palatal growth, interaction with rugae signalling centres and the maintenance of a neural stem cell niche [29]. Sox4 is situated at the nodal point where it can integrate several developmental pathways critical for secondary palate development, such as the TGFβ, and Wnt and Hippo signalling pathways [30]. Goldsworthy et al. [30]. have shown that 60% of the offspring from a cross between a mouse strain bearing a Sox4 mutation, and another bearing a Sox4 deletion, exhibit cleft palate [30]. They have also demonstrated that differential Sox4 expression during development of the secondary palate is regulated by DNA methylation, thereby making this gene a potential epigenetic target for environmental factors contributing to orofacial clefts [30]. Moreover, SOX4 is known to be overexpressed in a wide variety of human tumours, confirming an important role for this encoded protein in cell proliferation, differentiation, and apoptosis [31]. It is worth noting that SNPs within, and surrounding, the SOX4 gene were not associated with the risk of nsCL/P in our GWAS (results not shown).
The results of sequencing analysis may also provide indirect evidence that SOX4 is a possible cleft-susceptibility gene at the 6p22.3 locus. We did not find any potentially pathogenic mutations in the selected CDKAL1 exons in our 55 patients with nsCL/P. We detected only a rare missense variant (rs111739077, p.Arg39Gln) and six common polymorphisms located in the intronic sequences. The allele frequencies of all the identified variants did not differ among our Polish nsCL/P cases and individuals of European ancestry from the 1000 Genomes Project. In the large non-Finnish European population of ExAC the missense p.Arg39Gln variant has an allele frequency of 0.007, indicating that is too common to be causative [32]. In addition, eight CDKAL1 missense variants tested with the use of the SNP array platform were not detected in our nsCL/P cases or the healthy individuals.
The similar CDKAL1 association patterns between nsCL/P and T2DM might be due to the role of Sox4 in the development of endocrine pancreas [33]. Mice homozygous for a null mutation of Sox4 exhibit disturbed pancreatic bud formation and differentiation of endocrine cells [30]. Sox4 mutations in the adult mouse result in impaired glucose tolerance and insulin secretory defect [30]. In addition, it has been shown that increased SOX4 expression in human pancreatic islets correlates with reduced glucose-induced insulin secretion, which is a hallmark of T2DM [34]. However, it cannot be excluded that diabetes risk SNPs located in the fifth intron of CDKAL1 affect the function of the CDKAL1 gene itself. Mouse model studies revealed that Cdkal1 encodes a methylthiotransferase that modifies tRNALys to enhance translational fidelity of the proinsulin transcript [35]. Moreover, Cdkal1 modulates whole-body glucose metabolism in a bidirectional manner, since its lack enhances insulin sensitivity in various tissues and impairs insulin secretion in pancreatic β-cells [36]. It is interesting to note that orofacial clefts are one of the major malformations among offspring of women with diabetes [37, 38]. There are several mechanisms that could underline an association between maternal diabetes and congenital anomalies, including a teratogenic effect of hyperglycaemia or hyperinsulinemia [39, 40]. In addition, there is a possibility that the teratogenic effect of maternal diabetes might be a result of genetic factors related to diabetes-susceptibility genes [39].
The present study, despite its apparent strengths such as a homogenous study cohort recruited from a single ethnic group and confirmation of the significant results in the independent validation sample, has some limitations. These include the relatively small sample size, an association analysis focused only on the identification of common risk variants and the lack of information on maternal smoking and folate status during pregnancy, factors that may contribute to the aetiology of nsCL/P [41, 42]. Other weak points in the present research are sequencing analysis limited to the selected CDKAL1 coding exons and a lack of information about the functional significance of the identified risk SNPs. In addition, the most significant and positively replicated SNPs identified in our GWAS did not reach the threshold of genome-wide significance. Therefore, future studies will need to be undertaken to confirm our results in other populations and to narrow the candidate region to a smaller collection of SNPs. These could be further tested in functional assays such as in vitro analysis of cis-acting CDKAL1 variants on the SOX4 expression levels. It should be noted that the considerable proportion of the GWAS results with a borderline genome-wide significance represent replicable and possibly true SNP–disease associations [43].
The findings of this study suggest that chromosomal region 6p22.3 might be a novel susceptibility locus for nsCL/P. The location of the risk SNPs within the CDKAL1 intronic sequence comprising enhancer elements predicted to regulate the SOX4 transcription levels suggest that SOX4, rather than CDKAL1, is a potential candidate gene for this common craniofacial anomaly. Moreover, in contrast to CDKAL1, there is biological evidence supporting the role of SOX4 during palatogenesis.
References
Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–9.
MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E, et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 2017;45:D896–D901.
Tak YG, Farnham PJ. Making sense of GWAS: using epigenomics and genome engineering to understand the functional relevance of SNPs in non-coding regions of the human genome. Epigenetics Chromatin. 2015;8:57.
Dixon MJ, Marazita ML, Beaty TH, Murray JC. Cleft lip and palate: understanding genetic and environmental influences. Nat Rev Genet. 2011;12:167–78.
Birnbaum S, Ludwig KU, Reutter H, Herms S, Steffens M, Rubini M, et al. Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nat Genet. 2009;41:473–7.
Mangold E, Ludwig KU, Birnbaum S, Baluardo C, Ferrian M, Herms S, et al. Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate. Nat Genet. 2010;42:24–6.
Grant SF, Wang K, Zhang H, Glaberson W, Annaiah K, Kim CE, et al. A genome-wide association study identifies a locus for nonsyndromic cleft lip with or without cleft palate on 8q24. J Pediatr. 2009;155:909–13.
Beaty TH, Murray JC, Marazita ML, Munger RG, Ruczinski I, Hetmanski JB, et al. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat Genet. 2010;42:525–9.
Sun Y, Huang Y, Yin A, Pan Y, Wang Y, Wang C, et al. Genome-wide association study identifies a new susceptibility locus for cleft lip with or without a cleft palate. Nat Commun. 2015;6:6414.
Wolf ZT, Brand HA, Shaffer JR, Leslie EJ, Arzi B, Willet CE, et al. Genome-wide association studies in dogs and humans identify ADAMTS20 as a risk variant for cleft lip and palate. PLoS Genet. 2015;11:e1005059.
Leslie EJ, Carlson JC, Shaffer JR, Feingold E, Wehby G, Laurie CA, et al. A multi-ethnic genome-wide association study identifies novel loci for non-syndromic cleft lip with or without cleft palate on 2p24.2, 17q23 and 19q13. Hum Mol Genet. 2016;25:2862–72.
Uslu VV, Petretich M, Ruf S, Langenfeld K, Fonseca NA, Marioni JC, et al. Long-range enhancers regulating Myc expression are required for normal facial morphogenesis. Nat Genet. 2014;46:753–8.
Steinthorsdottir V, Thorleifsson G, Reynisdottir I, Benediktsson R, Jonsdottir T, Walters GB, et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet. 2007;39:770–5.
Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research, Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–6.
Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–41.
Peng F, Hu D, Gu C, Li X, Li Y, Jia N, et al. The relationship between five widely-evaluated variants in CDKN2A/B and CDKAL1 genes and the risk of type 2 diabetes: a meta-analysis. Gene. 2013;531:435–43.
Ragvin A, Moro E, Fredman D, Navratilova P, Drivenes Ø, Engström PG, et al. Long-range gene regulation links genomic type 2 diabetes and obesity risk regions to HHEX, SOX4, and IRX3. Proc Natl Acad Sci USA. 2010;107:775–80.
Juriloff DM, Harris MJ. Mouse genetic models of cleft lip with or without cleft palate. Birth Defects Res A Clin Mol Teratol. 2008;82:63–77.
Mostowska A, Hozyasz KK, Wójcicki P, Biedziak B, Wesoły J, Sowińska A, et al. Searching for new genes and loci involved in cleft lip and palate in the Polish population - genome-wide association study. J Med Sci. 2014;83:265–8.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75.
NCI-NHGRI Working Group on Replication in Association Studies, Chanock SJ, Manolio T, Boehnke M, Boerwinkle E, Hunter DJ, et al. Replicating genotype-phenotype associations. Nature. 2007;447:655–60.
Beaty TH, Marazita ML, Leslie EJ. Genetic factors influencing risk to orofacial clefts: today’s challenges and tomorrow’s opportunities. F1000Res. 2016;5:2800.
Li YY, Wang LS, Lu XZ, Yang ZJ, Wang XM, Zhou CW, et al. CDKAL1 gene rs7756992 A/G polymorphism and type 2 diabetes mellitus: a meta-analysis of 62,567 subjects. Sci Rep. 2013;3:3131.
Schilham MW, Moerer P, Cumano A, Clevers HC. Sox-4 facilitates thymocyte differentiation. Eur J Immunol. 1997;27:1292–5.
Cheung M, Abu-Elmagd M, Clevers H, Scotting PJ. Roles of Sox4 in central nervous system development. Brain Res Mol Brain Res. 2000;79:180–91.
Bhattaram P, Penzo-Méndez A, Sock E, Colmenares C, Kaneko KJ, Vassilev A, et al. Organogenesis relies on SoxC transcription factors for the survival of neural and mesenchymal progenitors. Nat Commun. 2010;1:9.
Lefebvre V, Bhattaram P. SOXC genes and the control of skeletogenesis. Curr Osteoporos Rep. 2016;14:32–8.
Hong CS, Saint-Jeannet JP. Sox proteins and neural crest development. Semin Cell Dev Biol. 2005;16:694–703.
Seelan RS, Mukhopadhyay P, Warner DR, Webb CL, Pisano M, Greene RM. Epigenetic regulation of Sox4 during palate development. Epigenomics. 2013;5:131–46.
Goldsworthy M, Hugill A, Freeman H, Horner E, Shimomura K, Bogani D, et al. Role of the transcription factor sox4 in insulin secretion and impaired glucose tolerance. Diabetes. 2008;57: 2234–44.
Vervoort SJ, van Boxtel R, Coffer PJ. The role of SRY-related HMG box transcription factor 4 (SOX4) in tumorigenesis and metastasis: friend or foe? Oncogene. 2013;32:3397–409.
Kobayashi Y, Yang S, Nykamp K, Garcia J, Lincoln SE, Topper SE. Pathogenic variant burden in the ExAC database: an empirical approach to evaluating population data for clinical variant interpretation. Genome Med. 2017;9:13.
Wilson ME, Yang KY, Kalousova A, Lau J, Kosaka Y, Lynn FC, et al. The HMG box transcription factor Sox4 contributes to the development of the endocrine pancreas. Diabetes. 2005;54: 3402–9.
Xu EE, Sasaki S, Speckmann T, Nian C, Lynn FC. SOX4 allows facultative β-cell proliferation through repression of Cdkn1a. Diabetes. 2017;66:2213–9.
Ohara-Imaizumi M, Yoshida M, Aoyagi K, Saito T, Okamura T, Takenaka H, et al. Deletion of CDKAL1 affects mitochondrial ATP generation and first-phase insulin exocytosis. PLoS ONE. 2010;5:e15553.
Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316: 1341–5.
Spilson SV, Kim HJ, Chung KC. Association between maternal diabetes mellitus and newborn oral cleft. Ann Plast Surg. 2001;47:477–81.
Negrato CA, Mattar R, Gomes MB. Adverse pregnancy outcomes in women with diabetes. Diabetol Metab Syndr. 2012;4:41.
Mills JL. Malformations in infants of diabetic mothers. Birth Defects Res A Clin Mol Teratol. 2010;88:769–78.
Kitzmiller JL, Wallerstein R, Correa A, Kwan S. Preconception care for women with diabetes and prevention of major congenital malformations. Birth Defects Res A Clin Mol Teratol. 2010;88: 791–803.
Wehby GL, Murray JC. Folic acid and orofacial clefts: a review of the evidence. Oral Dis. 2010;16:11–9.
Hackshaw A, Rodeck C, Boniface S. Maternal smoking in pregnancy and birth defects: a systematic review based on 173 687 malformed cases and 11.7 million controls. Hum Reprod Update. 2011;17:589–604.
Panagiotou OA, Ioannidis JP, Genome-Wide Significance Project. What should the genome-wide significance threshold be? Empirical replication of borderline genetic associations. Int J Epidemiol. 2012;41:273–86.
Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7.
Acknowledgements
The study was supported by a Polish National Science Centre grant no. 2012/07/B/NZ2/00115. The technical assistance of Justyna Dąbrowska and Sylwia Matuszewska-Trojan is gratefully acknowledged. We also wish to acknowledge the editorial assistance of Professor Geoffrey Shaw.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Gaczkowska, A., Żukowski, K., Biedziak, B. et al. Association of CDKAL1 nucleotide variants with the risk of non-syndromic cleft lip with or without cleft palate. J Hum Genet 63, 397–406 (2018). https://doi.org/10.1038/s10038-017-0397-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-017-0397-4
This article is cited by
-
The genetic factors contributing to the risk of cleft lip-cleft palate and their clinical utility
Oral and Maxillofacial Surgery (2022)
-
Identification of novel susceptibility genes for non-syndromic cleft lip with or without cleft palate using NGS-based multigene panel testing
Molecular Genetics and Genomics (2022)