Abstract
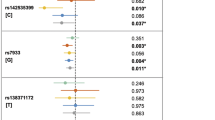
Non-syndromic cleft lip with or without cleft palate (NSCL/P) is a common polygenetic disease. Although genome-wide association studies (GWAS) identified NTN1 gene as a high-priority candidate of NSCL/P, the comprehensive genetic architecture of NTN1 weren’t yet known. Thus, this study aimed to determine full-scale genetic variants of NTN1 for NSCL/P in Chinese Han people. Initially, targeted sequencing of NTN1 gene was performed on 159 NSCL/P patients to identify susceptible single nucleotide polymorphisms (SNPs) associated with NSCL/P. Then, association analysis and burden analysis were separately used to validate the common variants and rare variants identified among large size of samples (1608 NSCL/P cases and 2255 controls). Additionally, NSCL/P subtype association analysis was applied to elucidate the etiology discrepancy of non-syndromic cleft lip with palate (NSCLP) and non-syndromic cleft lip only (NSCLO). Lastly, bioinformatics analysis was performed to annotate and prioritize candidate variants. We found 15 NSCL/P-associated SNPs including rs4791774 (P = 1.10E-08, OR = 1.467, 95% CI: 1.286~1.673) and rs9788972 (P = 1.28E-07, OR = 1.398, 95% CI : 1.235~1.584) originally detected by previous GWASs in Chinese Han ancestry. Four NSCLO risk-associated SNPs and eight specific NSCLP associated SNPs were found. Three SNPs (rs4791331, rs4791774 and rs9900753) were predicted to locate at regulatory region of NTN1. Our study validated the association between NTN1 gene and pathogenesis of NSCL/P and reinforced the hypothesis that NSCLP have a different etiology from NSCLO. We also identified three putative regulatory SNPs in NTN1 gene.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
Supplementary information is available at Journal of Human Genetics’s website.
References
Tanaka SA, Mahabir RC, Jupiter DC, Menezes JM. Updating the epidemiology of cleft lip with or without cleft palate. Plast Reconstr Surg. 2012;129:511e–8e.
Fan D, Wu S, Liu L, Xia Q, Tian G, Wang W, et al. Prevalence of non-syndromic orofacial clefts: based on 15,094,978 Chinese perinatal infants. Oncotarget. 2018;9:13981–90.
Sivertsen A, Wilcox A, Skjaerven R, Vindenes H, Abyholm F, Harville E, et al. Familial risk of oral clefts by morphological type and severity: population based cohort study of first degree relatives. BMJ. 2008;336:432–4.
Marazita ML. The evolution of human genetic studies of cleft lip and cleft palate. Annu Rev Genomics Hum Genet. 2012;13:263–83.
Mossey P, Little J, Munger R, Dixon M, Shaw W. Cleft lip and palate. Lancet 2009;374:1773–85.
Harville E, Wilcox A, Lie R, Vindenes H, Abyholm F. Cleft lip and palate versus cleft lip only: are they distinct defects? Am J Epidemiol. 2005;162:448–53.
Grosen D, Chevrier C, Skytthe A, Bille C, Mølsted K, Sivertsen A, et al. A cohort study of recurrence patterns among more than 54,000 relatives of oral cleft cases in Denmark: support for the multifactorial threshold model of inheritance. J Med Genet. 2010;47:162–8.
Beaty TH, Murray JC, Marazita ML, Munger RG, Ruczinski I, Hetmanski JB, et al. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat Genet. 2010;42:525–9.
Ludwig KU, Mangold E, Herms S, Nowak S, Reutter H, Paul A, et al. Genome-wide meta-analyses of nonsyndromic cleft lip with or without cleft palate identify six new risk loci. Nat Genet. 2012;44:968–71.
Beaty TH, Taub MA, Scott AF, Murray JC, Marazita ML, Schwender H, et al. Confirming genes influencing risk to cleft lip with/without cleft palate in a case-parent trio study. Hum Genet. 2013;132:771–81.
Leslie EJ, Carlson JC, Shaffer JR, Feingold E, Wehby G, Laurie CA, et al. A multi-ethnic genome-wide association study identifies novel loci for non-syndromic cleft lip with or without cleft palate on 2p24.2, 17q23 and 19q13. Hum Mol Genet 2016;25:2862–72.
Sun Y, Huang Y, Yin A, Pan Y, Wang Y, Wang C, et al. Genome-wide association study identifies a new susceptibility locus for cleft lip with or without a cleft palate. Nat Commun. 2015;6:6414.
Mukhopadhyay N, Feingold E, Moreno-Uribe L, Wehby G, Valencia-Ramirez LC, Muneton CPR, et al. Genome-wide association study of non-syndromic orofacial clefts in a multiethnic sample of families and controls identifies novel regions. Front Cell Dev Biol. 2021;9:621482.
Leslie EJ, Taub MA, Liu H, Steinberg KM, Koboldt DC, Zhang Q, et al. Identification of functional variants for cleft lip with or without cleft palate in or near PAX7, FGFR2, and NOG by targeted sequencing of GWAS loci. Am J Hum Genet. 2015;96:397–411.
Li D, Zhu G, Lou S, Ma L, Zhang C, Pan Y, et al. The functional variant of NTN1 contributes to the risk of nonsyndromic cleft lip with or without cleft palate. Eur J Hum Genet. 2020;28:453–60.
Tam V, Patel N, Turcotte M, Bosse Y, Pare G, Meyre D. Benefits and limitations of genome-wide association studies. Nat Rev Genet. 2019;20:467–84.
Bonnefond A, Froguel P. Rare and common genetic events in type 2 diabetes: what should biologists know? Cell Metab. 2015;21:357–68.
Sazonovs A, Barrett JC. Rare-variant studies to complement genome-wide association studies. Annu Rev Genomics Hum Genet. 2018;19:97–112.
International HapMap C. A haplotype map of the human genome. Nature. 2005;437:1299–320.
Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–8.
Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 2012;337:1190–5.
Thieme F, Ludwig KU. The role of noncoding genetic variation in isolated orofacial clefts. J Dent Res. 2017;96:1238–47.
Huang L, Jia Z, Shi Y, Du Q, Shi J, Wang Z, et al. Genetic factors define CPO and CLO subtypes of nonsyndromicorofacial cleft. PLOS Genet. 2019;15:e1008357.
Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–60.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics.2009;25:2078–9.
Yang J, Bakshi A, Zhu Z, Hemani G, Vinkhuyzen A, Lee S, et al. Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat Genet. 2015;47:1114–20.
Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013;Chapter 7:Unit7 20. https://doi.org/10.1002/0471142905.hg0720s76.
Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–4.
Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–6.
Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47:D886–D94.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75.
Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–7.
Quan C, Ping J, Lu H, Zhou G, Lu Y. 3DSNP 2.0: update and expansion of the noncoding genomic variant annotation database. Nucleic Acids Res. 2022;50:D950–D5.
Ward L, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–4.
Kou I, Otomo N, Takeda K, Momozawa Y, Lu HF, Kubo M, et al. Genome-wide association study identifies 14 previously unreported susceptibility loci for adolescent idiopathic scoliosis in Japanese. Nat Commun. 2019;10:3685.
Barbeira AN, Bonazzola R, Gamazon ER, Liang Y, Park Y, Kim-Hellmuth S, et al. Exploiting the GTEx resources to decipher the mechanisms at GWAS loci. Genome Biol. 2021;22:49.
Chen S, Jia Z, Cai M, Ye M, Wu D, Wan T, et al. viaSP1-mediated upregulation of long noncoding RNA ZFAS1 involved in non-syndromic cleft lip and palate inactivating WNT/β-catenin signaling pathway. Front Cell Dev Biol. 2021;9:662780.
Carlson J, Standley J, Petrin A, Shaffer J, Butali A, Buxó C, et al. Identification of 16q21 as a modifier of nonsyndromic orofacial cleft phenotypes. Genet Epidemiol. 2017;41:887–97.
Yin B, Shi JY, Lin YS, Shi B, Jia ZL. SNPs at TP63 gene was specifically associated with right-side cleft lip in Han Chinese population. Oral Dis. 2021;27:559–66.
Çalışkan M, Manduchi E, Rao H, Segert J, Beltrame M, Trizzino M, et al. Genetic and epigenetic fine mapping of complex trait associated loci in the human. Liver 2019;105:89–107.
Blanco-Gómez A, Castillo-Lluva S, Del Mar Sáez-Freire M, Hontecillas-Prieto L, Mao J, Castellanos-Martín A, et al. Missing heritability of complex diseases: enlightenment by genetic variants from intermediate phenotypes. Bioessays. 2016;38:664–73.
Field A, Adelman K. Evaluating enhancer function and transcription. Annu Rev Biochem. 2020;89:213–34.
Cramer P. Organization and regulation of gene transcription. Nature 2019;573:45–54.
Edwards S, Beesley J, French J, Dunning AM. Beyond GWASs: illuminating the dark road from association to function. Am J Hum Genet. 2013;93:779–97.
Guo Q, Li D, Meng X, Liu T, Shi J, Hao Y, et al. Association between PAX7 and NTN1 gene polymorphisms and nonsyndromic orofacial clefts in a northern Chinese population. Med (Baltim). 2017;96:e6724.
Xu Y, Xie B, Shi J, Li J, Zhou C, Lu W, et al. Distinct expression of miR-378 in nonsyndromic cleft lip and/or cleft palate: a cogitation of skewed sex ratio in prevalence. Cleft Palate Craniofac J. 2021;58:61–71.
Acknowledgements
The authors thank all the participants provided samples and friends donated helpful comments in this study.
Funding
This project was supported by the National Science Funds of China (No.82170919 and No. 81600849) and the Major frontier issues of the application foundation project of Sichuan Science and Technology Department (2020YJ0211).
Author information
Authors and Affiliations
Contributions
HXT Conceptualization, Methodology, Investigation, Writing-Original Draft. YXYang validation, Investigation, Writing-original draft, BS Validation, Software, Writing-Review/Editing, ZLJ Conceptualization, Methodology, Software, Supervision, Project administration, Funding acquisition, Writing-Review/Editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tao, HX., Yang, YX., Shi, B. et al. Identification of putative regulatory single-nucleotide variants in NTN1 gene associated with NSCL/P. J Hum Genet 68, 491–497 (2023). https://doi.org/10.1038/s10038-023-01137-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-023-01137-1