Abstract
Previously, we communicated that rapid and efficient purification of carbohydrates can be achieved by employing sugar-specific chemical ligation with hydrazide-functionalized polymer beads; we termed this process ‘glycoblotting’. The polymer beads are designed to recover carbohydrates by an imine exchange reaction in which hydrazone bonds between the beads and carbohydrates are transferred to oxime bonds between the aminooxy probe and the carbohydrates. To apply our concept to reductive amination with common fluorescent dyes, such as 2-aminobenzamide, the method for releasing the carbohydrates from the beads was examined, and we found that heating the beads with several percentages of acetic acid was efficient. Additionally, we obtained fundamental data on our novel method, such as the recovery ratio, the quantitative capability and the reproducibility. From the results, we concluded that rapid and accurate glycan analysis can be achieved with this novel method. Overall, these novel technologies represent a significant advance toward more efficient glycan analyses, especially by using high-performance liquid chromatography.
Similar content being viewed by others
Introduction
Glycosylation is a common post-translational modification of proteins. Most proteins are linked to glycans1, 2 which adjust the functions of these proteins for specific purposes by changing the protein folding, targeting, ligand recognition, binding, stability, immunogenicity and biological activity.3, 4, 5 Thus, glycans are now attracting much attention for a variety of biotechnology applications, including biomarker discovery and post-translational modification of therapeutic proteins.6, 7, 8, 9 It is well known that glycoproteins could be good tumor markers. For example, α-fetoprotein, prostate-specific antigen and carcinoembryonic antigen, which are cancer markers that are widely used in clinical applications, are types of glyco-markers.10, 11, 12, 13, 14, 15 Glycans also have important roles in therapeutic proteins. Accordingly, the evaluation of glycans as a quality control parameter for glycoprotein-based drugs is extremely important. This is particularly true in the field of antibody-based drugs; it has been reported that the glycan structure determines the antibody-dependent cellular cytotoxicity activity of such drugs, thus the structural analysis of the glycan is important.16, 17
Accordingly, techniques for analyzing glycan structure in a rapid, simple and precise manner have recently become very desirable. Glycan analyses are currently conducted using a wide variety of methods, including high-performance liquid chromatography (HPLC), nuclear magnetic resonance spectroscopy, capillary electrophoresis and mass analysis, among other techniques. To analyze glycans with these techniques, it is necessary to first isolate and purify the glycan from impurities, such as proteins, peptides, lipids and nucleic acids, contained in biological samples. Although HPLC and capillary electrophoresis have widely been adopted by virtue of their excellent resolution, good reproducibility, good quantification and high sensitivity, it is necessary to first label the reducing end of the glycan to obtain high sensitivity, and this is typically carried out by reductive amination. However, such purification and labeling of the glycan requires time and a significant number of process steps, so it is difficult to prepare a large amount of sample at one time.
Recently, Royle et al.18 have reported a method for glycan purification and labeling with 2-aminobenzamide (2-AB), which is one of the most widely used labels for HPLC analyses, with their in-gel block method. Their method is based on a 96-well plate format and enables high-throughput processing. However, their method is limited to a pretreatment method to release glycan from the glycoprotein. Therefore, their method does not accommodate samples that have been pretreated by other methods, such as trypsin treatment or hydrazinolysis.
Previously, we communicated that the rapid and efficient purification of glycans can be achieved by employing sugar-specific chemical ligation with hydrazide-functionalized polymers beads (termed ‘glycoblotting’19). With this technology, impurities such as salts, peptides, proteins and detergents can be completely removed. The polymer beads are designed to recover glycans by an imine exchange reaction in which hydrazone bonds between the beads and carbohydrates are transferred to oxime bonds between an aminooxy probe and the carbohydrate. Although this method has a great advantage in rapid glycan purification, this kind of labeling is limited in that the labeling of glycans with common fluorescent dyes, such as 2-AB,20, 21 2-aminobenzoic acid (2-AA),20 and 2-aminopyridine (2-AP),22, 23 which are useful for HPLC analysis, was not used in our previous technology. Therefore, it is impossible to compare the results obtained using ‘glycoblotting’ to individually accumulated data or existing databases, and thousands of hours and a great deal of effort would be required to obtain new data. Thus, our technology was further applied to the reductive amination with 2-AB, 2-AA and 2-AP. With the method described here, glycans are readily purified and labeled with amino compounds in the same reaction tube. Because our novel method is simple and does not require any special skills, rapid and accurate glycan analysis is readily provided. This work demonstrates that our novel technology is extremely useful as a pretreatment method for HPLC and liquid chromatography-mass spectrometry (LC-MS) analyses.
Experimental procedure
Materials
Bovine immunoglobulin (Ig)G, trypsin and sodium cyanoborohydride were obtained from Sigma-Aldrich (St Louis, MO, USA). Ammonium bicarbonate, dithiothreitol, 2-iodoacetamide, hydrochloric acid (HCl), acetic acid, acetonitrile (ACN), guanidine, triethylamine, methanol, acetic anhydride, 2-AB, dimethylsulfoxide, 2-AP, dimethylamineborane, formic acid and ammonia solution were purchased from Wako Pure Chemical Industries (Osaka, Japan). N-glycosidase F (PNGase F) was obtained from Roche Applied Science (Penzberg, Germany). NA2 and 2-AB-labeled NA2 were obtained from ProZyme (Hayword, CA, USA), and maltoheptaose was obtained from Seikagaku Biobusiness Corporation (Tokyo, Japan).
Synthesis
Preparation of a hydrazide-functionalized glycoblotting polymer (hydrazide beads)
The polymer synthesis was performed according to the method of Furukawa et al. using suspension polymerization.19 The beads are commercially available from Sumitomo Bakelite (Tokyo, Japan) as BlotGlyco.
Sample preparation
Preparation of crude glycan mixtures from model glycoprotein
Bovine IgG (1 mg) was added to 50 μl of 100 mM ammonium bicarbonate and 5 μl of 120 mM dithiothreitol and incubated at 60 °C for 0.5 h. Then, 10 μl of 123 mM 2-iodoacetamide in water was added, and the mixture was allowed to stand at ambient temperature for 1 h. Subsequently, 10 μl (400 U) of trypsin dissolved in 0.1 M HCl was added and incubated at 37 °C for 1 h, followed by heat denaturation at 90 °C for 5 min. Finally, 5 U of PNGase F was added and incubated at 37 °C for 12 h. The total volume was brought to 100 μl with water.
To compare our method to ethanol precipitation, IgG was pretreated by only using PNGase F (not containing protein fragmentation treatment). IgG (1 mg) was added to 50 μl of 100 mM ammonium bicarbonate and 5 U of PNGase F, followed by incubation at 37 °C for 12 h. The total volume was brought to 100 μl with water.
Capture of glycans by the hydrazide beads
In a disposable filter tube, 20 μl of the sample solution (including glycans) and 180 μl of 2% acetic acid/ACN were added to 5 mg of the hydrazide beads and incubated at 80 °C for 0.5 h while keeping the tube lid open. Then, the beads were washed with 2 M guanidine, water, 1% triethylamine/methanol and methanol to remove nonspecifically bound impurities. After washing, the beads were treated with 10% acetic anhydride/methanol for 0.5 h at ambient temperature to quench the hydrazide groups. The beads were then washed with methanol and water.
Release of glycans from the hydrazide beads
In the filter tube, 20 μl of water and 180 μl of ACN with various concentrations of acetic acid were added to the beads and incubated at 70 °C for 1.5 h, keeping the lid open to release the glycans attached to the beads.
2-AB labeling of the released glycans
After releasing the glycans from the beads, 50 μl of the labeling reagent (0.35 M 2-AB, 1 M sodium cyanoborohydride in dimethylsulfoxide-acetic acid (70:30, v/v)) was added to the beads in the filter tube and incubated at 60 °C for 2 h while keeping the tube lid open.20 The 2-AB-labeled glycans were then separated from the excess reagent using silica gel columns as follows. The columns were conditioned with 200 μl of water and 200 μl of ACN. The 2-AB-labeled glycan solutions were diluted 20 times with ACN and passed through the columns. Then, the columns were washed with 400 μl of ACN three times and 400 μl of 95% ACN/water three times, and the 2-AB-labeled glycans were eluted with 50 μl of water.
For an optimization study of glycan release, the 2-AB labeling was performed in a 1.5-ml reaction tube, rather than on beads. Model glycans (bovine IgG containing ∼2 nmol of N-type glycans) were captured by the hydrazide beads as described above. After sequential washing and capping of the remaining hydrazide groups, the glycans immobilized on the beads were released by treatment with ACN solutions containing various concentrations of acetic acid (0, 0.1, 1, 2, 5, 10 and 20% acetic acid). To the beads, 50 μl of water was added and recovered by centrifugation. The recovered solution contained glycans released from the beads and was evaporated to dryness using a vacuum centrifugal evaporator. The recovered solution was labeled with 2-AB.
2-AP labeling of the released glycans
After releasing the glycans from the beads, 30 μl of the labeling reagent (552 mg of 2-AP dissolved in 200 μl of acetic acid) was added to the beads in the filter tube and heated to 90 °C for 1 h, keeping the tube lid open.23, 24, 25 After completion of the coupling reaction, 110 μl of reducing reagent was added. The reducing agent was freshly prepared by dissolving 200 mg of dimethylamineborane complex in 50 μl of H2O and 80 μl of acetic acid. The pyridylamino derivatives of glycans were purified by silica gel columns as follows. The columns were conditioned with 200 μl of water and 200 μl of ACN. The 2-AP-labeled glycan solutions were diluted 10 times with ACN and passed through the columns. Then, the columns were washed with 400 μl of ACN three times and 400 μl of 95% ACN/water three times, and the 2-AP-labeled glycans were eluted with 50 μl of water.
2-AA labeling of the released glycans
After releasing the glycans from the beads, 50 μl of labeling reagent (0.35 M 2-AA, 1 M sodium cyanoborohydride in dimethylsulfoxide-acetic acid (70:30, v/v)) was added to the beads in the filter tube and incubated at 60 °C for 2 h (keeping the tube lid open), and the 2-AA-labeled glycans were then separated from excess reagent with silica gel columns as follows.20 The columns were conditioned with 200 μl of water and 200 μl of ACN. The 2-AA-labeled glycan solutions were diluted 20 times with ACN and passed through the columns. Then, the columns were washed with 400 μl of ACN three times and 400 μl of 95% ACN/water three times, and the 2-AA-labeled glycans were eluted with 50 μl of water.
Purification of bovine IgG glycans by ethanol precipitation
In this step, 200 μl of chilled ethanol was added to 60 μl of the deglycosylated bovine IgG solution (without protein fragmentation) and centrifuged (10 000 × g) for 10 min. The supernatant was decanted and evaporated to dryness using a vacuum centrifugal evaporator. The recovered glycans were dissolved in 60 μl of pure water.
Measurements
HPLC systems and conditions
HPLC separations were performed on a 2695 Alliance separation module (Waters, Milford, MA, USA) equipped with a Waters temperature control module and a Waters 4775 fluorescence detector. All HPLC systems were controlled via Waters Empower software. The HPLC analysis was performed using a TSK-GEL Amide-80 column (4.6 mm × 250 mm, TOSOH, Tokyo, Japan) with a gradient of 50 mM ammonium formate adjusted to pH 4.4 (buffer A) and ACN (buffer B) at 30 °C. A linear gradient was run from 20 to 58% buffer A over 150 min (at a flow rate of 0.4 ml min−1). Additionally, HPLC was performed using an Asahi Shodex NH2P-50 4E column (4.6 mm × 250 mm, Showa Denko, Tokyo, Japan) with a gradient of 2% acetic acid in ACN (buffer A) and 5% acetic acid in water containing 3% triethylamine (buffer B) at 40 °C. The column was initially equilibrated and eluted with 30% buffer B for 2 min, at which point the concentration of buffer B was increased to 95% over 80 min (at a flow rate of 1.0 ml min−1). Detection was performed by fluorometry at λex=330 nm and λem=420 nm (for 2-AB-labeled glycans), λex=360 nm and λem=425 nm (for 2-AA-labeled glycans), and λex=320 nm and λem=400 nm (for 2-AP-labeled glycans).
Matrix-assisted laser desorption ionisation time-of-flight MS (MALDI-TOF MS)
All measurements were performed using an Autoflex III TOF/TOF mass spectrometer equipped with a reflector and controlled by the FlexControl 2.1 software package (Bruker Daltonics GmbsH, Bremen, Germany). In MALDI-TOF MS reflector mode, ions generated by a pulsed ultraviolet laser beam (nitrogen laser, l=337 nm, 5 Hz) were accelerated to a kinetic energy of 23.5 keV. Masses were automatically annotated with the FlexAnalysis 2.0 (Bruker Daltonik, Bremen, Germany) software package. External calibration of MALDI mass spectra was carried out using singly charged monoisotopic peaks of a mixture of human angiotensin II (m/z 1046.542), bombesin (m/z 1619.823), adrenocorticotropin (18–39) (m/z 2465.199) and somatostatin 28 (m/z 3147.472). All measurements were performed as follows: 1 μl of the sample solution was mixed with 1 μl of 2,5-dihydroxybenzoic acid (DHB) (10 mg ml−1 in 30% ACN), and 1 μl of the resulting mixture was subjected to MALDI-TOF mass analysis.
LC-MS systems and conditions
HPLC separations were performed on an LC-MS-IT-TOF (Shimadzu Corporation, Kyoto, Japan) equipped with a LC-3AD solution pump, a DGU-20A3 degasser, an SIL-30AC autosampler, a CTO-20AC column oven and RF-20AXS fluorescence detectors. All HPLC systems were controlled via a CBM-20A system controller. HPLC was performed using an Aquity UPLC BEH Glycan 1.7 μm (2.1 mm × 150 mm, Waters) with a gradient of 0.5% formic acid aqueous solution (buffer A) and 0.5% formic acid ACN solution (buffer B) at 40 °C. A linear gradient was run from 27 to 40% buffer A over 25 min (at a flow rate of 0.4 ml min−1). All MS analyses were performed using electrospray ionization in the positive ion mode with a probe voltage of 4.5 kV, a CDL temperature of 200 °C, a nebulizing gas flow of 1.5 l min−1, a drying gas pressure of 0.1 MPa and a m/z scan range of 500–2500. External calibration of the MALDI mass spectra was carried out using sodium trifluoroacetate.
Results and Discussion
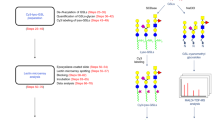
The general strategy employed in this study is outlined in Figure 1. The fundamental concept is based on previous reports.19, 26, 27, 28, 29, 30, 31, 32 Glycoproteins were purified and labeled with the hydrazide beads. At first, glycoproteins are pretreated to deglycosylate them by using enzymatic or chemical methods, and the released glycans are captured by the hydrazide beads through a hydrazone bond. The impurities contained in the glycan samples are completely removed by the subsequent washing steps, and the remaining hydrazide groups are quenched. Then, the glycans immobilized on the beads are released and labeled with the appropriate tags by reductive amination. This series of manipulations is performed in the same tube.
General strategy for the purifying and labeling of the glycans using the hydrazide beads. (a) Pretreatment of glycoproteins. (b) Capture of released glycans and removal of impurities. (c) Quenching of hydrazide groups. (d) Release of the glycans and labeling of the released glycans. All manipulations are performed in the same tube. A full color version of this figure is available at Polymer Journal online.
First, the method of glycan release was examined. The releasing reaction was performed while keeping the lid of the spin tube open to dry the beads. This served to raise the reaction yield and dry the released glycans for the subsequent labeling reaction. By releasing and drying the glycans in parallel, the operation time was reduced, and glycan labeling proceeded reproducibly. Figure 2 shows the efficiencies of the different concentrations of acetic acid for the release of the glycans from the hydrazide beads. Model glycans (bovine IgG) were treated with the hydrazide beads as described above. The 2-AB-labeled glycans were analyzed by NP-HPLC (Asahi Shodex NH2P-50 4E column) according to the method of Komoda et al.33 When the concentration of acetic acid was less than 1%, the releasing ratio decreased significantly. The ratio reached a maximum between 2 and 10% acetic acid and began to decrease at levels greater than 10%. In contrast, the loss of sialic acid residues occurred as the concentration of acetic acid was increased (data not shown). Consequently, we adopted 2% acetic acid as the reaction condition. Additionally, cleavage of sialic acid residues was observed to occur as the reaction temperature was raised. However, the time to dry the beads increased with decreasing reaction temperature (data not shown). As a result, we adopted the following reaction conditions for glycan release: 20 μl of water and 180 μl of 2% AcOH/ACN were added to the beads in the filter tube and incubated at 70 °C for 1.5 h, keeping the tube lid open. These reaction conditions accomplished the release of the glycans with a high recovery ratio (to be discussed below) and low sialic acid elimination (<5%, data not shown).
Efficiencies of different concentrations of acetic acid for releasing the glycans from the hydrazide beads. The immobilized glycans were released by reaction with various concentrations of acetic acid. After 2-aminobenzamide (2-AB) labeling, the released glycans were analyzed by HPLC. The vertical axis is the total peak area of 2-AB-labeled glycans, and the horizontal axis is the acetic acid concentration. A full color version of this figure is available at Polymer Journal online.
The feasibility of combining the hydrazide beads and 2-AB labeling was evaluated using bovine IgG. Obtained glycans were analyzed using HPLC and MALDI-TOF MS. HPLC analyses were performed using an NP-column (amide-80) according to the method of Royle et al.34 Glycan profiles obtained from HPLC analyses were calibrated against a 2-AB-labeled dextran ladder and assigned glucose unit (GU) values by fitting a polynomial distribution curve to allocate the glucose unit from retention times.34 Glycan structures were assigned using the database ‘Glycobase,’ which contains the experimental HPLC elution positions for 2-AB-labeled glycans expressed in the form of glucose unit values, as well as predicted products and MALDI-TOF MS results.35 Figure 3 shows the HPLC (A) and MALDI-TOF MS (B) profiles. Almost all molecular weights were detected as the sodium adduct ions (M+Na)+ in the MALDI spectrum. As expected, biantennary complex glycans with gal (0, 1, and 2) and core fucose were strongly detected by HPLC and MALDI-TOF MS. The glycans containing N-glycolyl sialic acid, which exist in bovine and porcine samples but not normally in human36, 37, 38 samples, were detected by HPLC analysis. These sialoglycans were not detected by MALDI-TOF MS analysis because losses of sialic acid occurred during the MALDI-TOF MS analysis. These results demonstrated that 2-AB-labeled glycans were obtained using our novel method and that purification and labeling of glycans could be performed in the same single vessel without complicated operations. This novel method only requires solution dispensing and centrifugation.
The 2-aminobenzamide-labeled glycans obtained by our novel method were detected by high-performance liquid chromatography (HPLC) and matrix-assisted laser desorption ionisation time-of-flight mass spectrometry (MALDI-TOF MS). (a) HPLC analysis was performed using an NP-column, and peaks were assigned glucose unit values by comparison with a standard dextran ladder. Only the major structures are shown in structural detail. (b) MALDI-TOF MS analysis was performed in reflection mode. All molecular weights represent monoisotopic masses of the respective [M+Na]+ ions of glycan species. Gal, galactose; GlcNAc, N-acetylglucosamine; Fuc, fucose; Man, mannose; NeuGc, N-glycolylneuraminic acid (N-glycolylsialic acid). A full color version of this figure is available at Polymer Journal online.
The method described here was compared with ethanol precipitation, which is an effective method for removing proteins from glycans. Bovine IgG deglycosylated only by PNGase F was purified using ethanol precipitation and our novel method and labeled with 2-AB as described above. The 2-AB-labeled glycans obtained were analyzed by HPLC using an NP-column (Shodex-NH2). Figure 4 shows the HPLC profiles. The ethanol precipitation method yielded the same glycan pattern and recovered the same amount of glycan as our method. This result indicates that the novel method presented here is comparable to ethanol precipitation, and no selective losses of particular types of glycans were associated with our novel method. Additionally, it is difficult to remove low-molecular-weight species, such as peptides and detergents, from glycans by ethanol precipitation due to the water solubility of these molecules. This is problematic because most glycoproteins require peptide fragmentation or denaturation by detergents to accomplish complete deglycosylation.39 In contrast, our novel method enables the removal of all impurities, including such peptides and detergents. Because glycans are captured on bead surfaces using the chemical reactivity of the glycans’ reducing ends, the method makes possible a complete wash to remove all impurities in glycan samples. This indicates that our novel method has substantial advantages in the purification and labeling of glycans included in every biological sample.
High-performance liquid chromatography profiles of the results of ethanol precipitation (a) and the novel method (b). The methods were compared using bovine immunoglobulin G deglycosylated only by PNGase F. (c) The amounts of glycan recovered from each method were also compared. The vertical axis is the total peak area of 2-aminobenzamide-labeled glycans, and the horizontal axis is the pretreatment method. A full color version of this figure is available at Polymer Journal online.
The question of the fraction of glycans recovered and labeled was addressed by two different experiments. First, commercially available 10 μM NA2 was treated with the hydrazide beads as described above, and 10 μl of the obtained 2-AB-labeled NA2 was analyzed by HPLC. In parallel, 10 pmol of commercially available 2-AB-labeled NA2 was also analyzed by HPLC as a glycan standard to calculate the recovery ratio. The recovery ratio was also confirmed using samples that contained impurities, such as salts, peptides and EDTA. The NA2 sample was spiked in bovine IgG solution pretreated with DTT, trypsin and PNGase F, and the mixed solution was hydrazide beads treated as described above. The recovery ratio of NA2 was estimated in the same way as that of the commercially available NA2. Because the IgG solution itself contained small amounts of NA2, the recovery ratio was calculated after determining the NA2 from the IgG glycan measurement. Table 1 shows the peak area and recovery ratio of NA2 obtained from each experiment. For the IgG-containing sample, the peak area of the recovered NA2 shown in Table 1 has been adjusted for the NA2 contained in the IgG itself. The glycan recovery ratio from the sample containing only pure glycan was 74.5%, whereas the ratio was 72% for the sample containing many impurities. These results demonstrated that the glycan recovery ratio of our novel method was ∼70%, regardless of the sample condition.
The quantitative capability of our novel method was confirmed using various concentrations of maltoheptaose solution (between 0.1 and 50 mM). Each maltoheptaose solution consisted of hydrazide beads and was treated as described above and was analyzed by HPLC using an NP-column (Amide-80). The correlation between the peak area and the concentration of maltoheptaose was verified. As shown in Figure 5, the peak area from maltoheptaose increased with rising concentration. The peak increased linearly from 0.1 μM to 5 mM; however, the recovery ratio gradually decreased. This is attributed to the fact that the amount of maltoheptaose was close to the amount of hydrazide groups possessed by the hydrazide beads. However, the results showed that our novel method enables quantitative analyses over a wide range of glycan concentrations (between 0.1 μM and 5 mM). Furthermore, quantitative capability was confirmed using solutions containing two kinds of glycan. Glycans of two different types were mixed in various ratios (100:1, 50:1, 10:1 and 1:1) and treated with the hydrazide beads as described above. The 2-AB-labeled glycans were detected using HPLC. Consequently, it was confirmed that the mixed glycans were captured and 2-AB labeled by the beads while maintaining their ratio in the original mixture (data not shown).
The correlation between peak area and concentration of maltoheptaose was verified. The vertical axis is the total peak area of 2-aminobenzamide-labeled maltoheptaose, and the horizontal axis is its concentration. (a) Overall view of results. (b) Close-up between 0.1 μM and 5 mM of maltoheptaose. (c) Close-up between 0.1 and 10 μM. A full color version of this figure is available at Polymer Journal online.
Using the hydrazide beads, glycans released from 1 μg of glycoprotein applied to the beads was always sufficient to obtain at least a glycan profile by HPLC. The reproducibility of the described technique was investigated by analysis of bovine IgG N-glycans in three separate experimental trials (n=3 for each trial). Each experiment included capturing glycans, capping hydrazide groups, releasing the immobilized glycans, and 2-AB labeling. The relative abundance of each glycan and the recovery amounts were compared. The within-assay coefficient of variation for glycan recovery varied from 2.9 to 3.8%, and the between-assay coefficient of variation was on the order of 7.0% (Figure 6a and Table 2). When considering the relative ratios, the within-assay coefficient of variation varied from 0.5 to 10.6%, and the between-assay coefficient of variation varied from 0.7 to 16.7%. Furthermore, both types of relative ratio CVs were lower (<10%) for glycan species whose relative proportion was >0.5% (Figures 6b and c and Table 3).
The reproducibility of the described technique was investigated by analysis of bovine immunoglobulin G N-glycans in three separate experimental trials (n=3 for each trial). (a) The recovery amount of 2-aminobenzamide (2-AB)-labeled IgG glycans on each assay was calculated and graphed. The vertical axis is the total peak area of 2-AB-labeled glycans. (b) The peak patterns within each assay were compared using the results from day 1. The vertical axis is the peak ratio of each glycan, and the horizontal axis is the number of peaks. (c) The peak patterns between assays were compared using the first result on each day. The vertical axis is the peak ratio of each glycan, and the horizontal axis is the number of peaks. A full color version of this figure is available at Polymer Journal online.
Purified and 2-AB-labeled glycans produced using our novel method were analyzed by LC-MS (LCMS-IT-TOF, Shimadzu Corporation). The total ion chromatogram exhibited a similar shape to that obtained with fluorescence detection (Figure 7). Impurities lacking a fluorescing substance, such as peptide and protein, were not detected. This indicated that all impurities included in the glycan samples were completely removed using our method. In particular, the isolation of glycans from peptide-included samples is difficult because the molecular weights of the peptides are similar to those of the glycans, and some peptides have a water affinity similar to that of the glycans. This result demonstrated that our novel method is useful to purify glycans from any sample.
Our novel method was applied to other general labeling processes, such as 2-AP and 2-AA labeling. Glycans from bovine IgG were captured using the hydrazide beads and released as described above. The obtained glycans were analyzed using HPLC (Asahi Shodex NH2P-50 4E column). Consequently, the glycans attached to each tag were clearly detected by HPLC. As noted in Figure 8, a pattern similar to that of the 2-AB-labeled IgG glycans was observed from the results using both tags. This indicates that our novel method works with labeling accomplished by reductive amination.
This paper has shown that intact glycans can be recovered and labeled with fluorescent tags, such as 2-AB, 2-AA and PA, with polymer beads possessing a high density of hydrazide groups. This technology offers a number of important advantages. First, the entire analysis (from the purification of the released glycans) can be performed in just 1 day without tedious operations. Additionally, our technology does not limit the method for glycan release because the technology is based on the chemoselective capturing of glycans, and all impurities are removed by the subsequent wash. This means that the method can accommodate a wide range of glycan samples, including various impurities such as salts, peptides and detergents. In addition, several samples can be processed in parallel. It can therefore be envisaged that large numbers of glycan samples could be investigated in a short time. We also developed a 96-well plate format protocol to accomplish high-throughput glycan purification and labeling. This system is amenable to automation and will make a significant contribution to the quality control of glycoprotein pharmaceuticals and large-scale biomarker discovery. A novel protocol focused on quality control for antibody drug processing is currently under development. Our hydrazide bead method holds the promise of becoming a standard method in the future owing to its high throughput capability, reproducibility and accuracy of glycan analysis.
References
Apweiler, R., Hermjakob, H. & Sharon, N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta. 1473, 4–8 (1999).
Ben-Dor, S., Esterman, N., Rubin, E. & Sharon, N. Biases and complex patterns in the residues flanking protein N-glycosylation sites. Glycobiology 14, 95–101 (2004).
Taylor, M. E. & Drickamer, K. Introduction to Glycobiology (Oxford University Press Inc., New York, 2003).
Helenius, A. & Aebi, M. Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73, 1019–1049 (2004).
Spiro, R. G. Role of N-linked polymannose oligosaccharides in targeting glycoproteins for endoplasmic reticulum-associated degradation. Cell Mol. Life Sci. 61, 1025–1041 (2004).
Allen, A. C., Harper, S. J. & Feehally, J. Galactosylation of N- and O-linked carbohydrate moieties of IgA1 and IgG in IgA nephropathy. Clin. Exp. Immunol. 100, 470–474 (1995).
Tomana, M., Matousovic, K., Julian, B. A., Radl, J., Konecny, K. & Mestecky, J. Galactose-deficient IgA1 in sera of IgA nephropathy patients is present in complexes with IgG. Kidney Int. 52, 509–516 (1997).
Aoyagi, Y. Carbohydrate-based measurements on alpha-fetoprotein in the early diagnosis of hepatocellular carcinoma. Glycoconj. J. 12, 194–199 (1995).
Seko, A., Kataoka, F., Aoki, D., Sakamoto, M., Nakamura, T., Hatae, M., Yonezawa, S. & Yamashita, K. N-acetylglucosamine 6-o-sulfotransferase-2 as a tumor marker for uterine cervical and corpus cancer. Glycoconj. J. 26, 1065–1073 (2009).
Abelev, G. I., Perova, S. D., Khramkova, N. I., Postnikova, Z. A. & Irlin, I. S. Production of embryonal alpha-globulin by transplantable mouse hepatomas. Transplantation 1, 174–180 (1963).
Egan, M. L., Pritchard, D. G., Todd, C. W. & Go, V. L. Isolation and immunochemical and chemical characterization of carcinoembryonic antigen-like substances in colon lavages of healthy individuals. Cancer Res. 37, 2638–2643 (1977).
Fritsche, R. & Mach, J. P. Isolation and characterization of carcinoembryonic antigen (CEA) extracted from normal human colon mucosa. Immunochemistry 14, 119–127 (1977).
Haynes, P. A., Gooley, A. A., Ferguson, M. A., Redmond, J. W. & Williams, K. L. Post-translational modifications of the Dictyostelium discoideum glycoprotein PsA. Glycosylphosphatidylinositol membrane anchor and composition of O-linked oligosaccharides. Eur. J. Biochem. 216, 729–737 (1993).
Nishio, K. Biomarkers for cancer molecular target therapy. Jpn. J. Clin. Oncol. 41, 95–100 (2010).
Thompson, J. A., Pande, H., Paxton, R. J., Shively, L., Padma, A., Simmer, R. L., Todd, C. W., Riggs, A. D. & Shively, J. E. Molecular cloning of a gene belonging to the carcinoembryonic antigen gene family and discussion of a domain model. Proc. Natl. Acad. Sci. USA 84, 2965–2969 (1987).
Shields, R. L., Lai, J., Keck, R., O’Connell, L. Y., Hong, K., Meng, Y. G., Weikert, S. H. & Presta, L. G. Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human Fcgamma RIII and antibody-dependent cellular toxicity. J. Biol. Chem. 277, 26733–26740 (2002).
Shinkawa, T., Nakamura, K., Yamane, N., Shoji-Hosaka, E., Kanda, Y., Sakurada, M., Uchida, K., Anazawa, H., Satoh, M., Yamasaki, M., Hanai, N. & Shitara, K. The absence of fucose but not the presence of galactose or bisecting N-acetylglucosamine of human IgG1 complex-type oligosaccharides shows the critical role of enhancing antibody-dependent cellular cytotoxicity. J. Biol. Chem. 278, 3466–3473 (2003).
Royle, L., Campbell, M. P., Radcliffe, C. M., White, D. M., Harvey, D. J., Abrahams, J. L., Kim, Y. G., Henry, G. W., Shadick, N. A., Weinblatt, M. E., Lee, D. M., Rudd, P. M. & Dwek, R. A. HPLC-based analysis of serum N-glycans on a 96-well plate platform with dedicated database software. Anal. Biochem. 376, 1–12 (2008).
Furukawa, J., Shinohara, Y., Kuramoto, H., Miura, Y., Shimaoka, H., Kurogochi, M., Nakano, M. & Nishimura, S.- I. Comprehensive approach to structural and functional glycomics based on chemoselective glycoblotting and sequential tag conversion. Anal. Chem. 80, 1094–1101 (2008).
Bigge, J. C., Patel, T. P., Bruce, J. A., Goulding, P. N., Charles, S. M. & Parekh, R. B. Nonselective and efficient fluorescent labeling of glycans using 2-amino benzamide and anthranilic acid. Anal. Biochem. 230, 229–238 (1995).
Townsend, R. R., Lipniunas, P. H., Bigge, C., Ventom, A. & Parekh, R. Multimode high-performance liquid chromatography of fluorescently labeled oligosaccharides from glycoproteins. Anal. Biochem. 239, 200–207 (1996).
Hase, S., Hara, S. & Matsushima, Y. Tagging of sugars with a fluorescent compound, 2-aminopyridine. J. Biochem. 85, 217–220 (1979).
Kondo, A., Suzuki, J., Kuraya, N., Hase, S., Kato, I. & Ikenaka, T. Improved method for fluorescence labeling of sugar chains with sialic acid residues. Agric. Biol. Chem. 54, 2169–2170 (1990).
Nakagawa, H., Kawamura, Y., Kato, K., Shimada, I., Arata, Y. & Takahashi, N. Identification of neutral and sialyl N-linked oligosaccharide structures from human serum glycoproteins using three kinds of high-performance liquid chromatography. Anal. Biochem. 226, 130–138 (1995).
Takahashi, N., Nakagawa, H., Fujikawa, K., Kawamura, Y. & Tomiya, N. Three-dimensional elution mapping of pyridylaminated N-linked neutral and sialyl oligosaccharides. Anal. Biochem. 226, 139–146 (1995).
Nishimura, S.- I., Niikura, K., Kurogochi, M., Matsushita, T., Fumoto, M., Hinou, H., Kamitani, R., Nakagawa, H., Deguchi, K., Miura, N., Monde, K. & Kondo, H. High-throughput protein glycomics: combined use of chemoselective glycoblotting and MALDI-TOF/TOF mass spectrometry. Angew. Chem. Int. Ed. Engl. 44, 91–96 (2005).
Nishimura, S.- I. Toward automated glycan analysis. Adv. Carbohydr. Chem. Biochem. 65, 211–264 (2011).
Niikura, K., Kamitani, R., Kurogochi, M., Uematsu, R., Shinohara, Y., Nakagawa, H., Deguchi, K., Monde, K., Kondo, H. & Nishimura, S.- I. Versatile glycoblotting nanoparticles for high-throughput protein glycomics. Chem. Eur. J. 11, 3825–3834 (2005).
Shimaoka, H., Kuramoto, H., Furukawa, J., Miura, Y., Kurogochi, M., Kita, Y., Hinou, H., Shinohara, Y. & Nishimura, S. One-pot solid-phase glycoblotting and probing by transoximization for high-throughput glycomics and glycoproteomics. Chem. Eur. J. 13, 1664–1673 (2007).
Shinohara, Y., Furukawa, J., Niikura, K., Miura, N. & Nishimura, S. Direct N-glycan profiling in the presence of tryptic peptides on MALDI-TOF by controlled ion enhancement and suppression upon glycan-selective derivatization. Anal. Chem. 76, 6989–6997 (2004).
Miura, Y., Shinohara, Y., Furukawa, J., Nagahori, N. & Nishimura, S.- I. Rapid and simple solid-phase esterification of sialic acid residues for quantitative glycomics by mass spectrometry. Chem. Eur. J. 13, 4797–4804 (2007).
Hirose, K., Amano, M., Hashimoto, R., Lee, Y. C. & Nishimura, S.- I. Insight into glycan diversity and evolutionary lineage based on comparative Avio-N-glycomics and sialic acid analysis of 88 egg whites of galloanserae. Biochemistry (Mosc.) 50, 4757–4774 (2011).
Kamoda, S., Nakano, M., Ishikawa, R., Suzuki, S. & Kakehi, K. Rapid and sensitive screening of N-glycans as 9-fluorenylmethyl derivatives by high-performance liquid chromatography: a method which can recover free oligosaccharides after analysis. J. Proteome Res. 4, 146–152 (2005).
Guile, G. R., Rudd, P. M., Wing, D. R., Prime, S. B. & Dwek, R. A. A rapid high-resolution high-performance liquid chromatographic method for separating glycan mixtures and analyzing oligosaccharide profiles. Anal. Biochem. 240, 210–226 (1996).
Campbell, M. P., Royle, L., Radcliffe, C. M., Dwek, R. A. & Rudd, P. M. GlycoBase and autoGU: tools for HPLC-based glycan analysis. Bioinformatics 24, 1214–1216 (2008).
Irie, A., Koyama, S., Kozutsumi, Y., Kawasaki, T. & Suzuki, A. The molecular basis for the absence of N-glycolylneuraminic acid in humans. J. Biol. Chem. 273, 15866–15871 (1998).
Chou, H. H., Takematsu, H., Diaz, S., Iber, J., Nickerson, E., Wright, K. L., Muchmore, E. A., Nelson, D. L., Warren, S. T. & Varki, A. A mutation in human CMP-sialic acid hydroxylase occurred after the Homo-Pan divergence. Proc. Natl. Acad. Sci. USA 95, 11751–11756 (1998).
Hayakawa, T., Satta, Y., Gagneux, P., Varki, A. & Takahata, N. Alu-mediated inactivation of the human CMP- N-acetylneuraminic acid hydroxylase gene. Proc. Natl. Acad. Sci. USA 98, 11399–11404 (2001).
Kita, Y., Miura, Y., Furukawa, J., Nakano, M., Shinohara, Y., Ohno, M., Takimoto, A. & Nishimura, S.- I. Quantitative glycomics of human whole serum glycoproteins based on the standardized protocol for liberating N-glycans. Mol. Cell Proteomics 6, 1437–1445 (2007).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Abe, M., Shimaoka, H., Fukushima, M. et al. A cross-linked polymer possessing a high density of hydrazide groups: high-throughput glycan purification and labeling for high-performance liquid chromatography analysis. Polym J 44, 269–277 (2012). https://doi.org/10.1038/pj.2011.125
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/pj.2011.125