Abstract
Background:
ERG rearrangements and PTEN (phosphatase and tensin homolog deleted on chromosome 10) loss are two of the most common genetic alterations in prostate cancer. However, there is still significant controversy regarding the order of events of these two changes during the carcinogenic process. We used immunohistochemistry (IHC) to determine ERG and PTEN status, and calculated the fraction of cases with homogeneous/heterogeneous ERG and PTEN staining in a given tumor.
Methods:
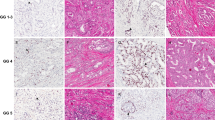
Using a single standard tissue section from the index tumor from radical prostatectomies (N=77), enriched for relatively high grade and stage tumors, we examined ERG and PTEN status by IHC. We determined whether ERG or PTEN staining was homogeneous (all tumor cells staining positive) or heterogeneous (focal tumor cell staining) in a given tumor focus.
Results:
Fifty-seven percent (N=44/77) of tumor foci showed ERG positivity, with 93% of these (N=41/44) cases showing homogeneous ERG staining in which all tumor cells stained positively. Fifty-three percent (N=41/77) of tumor foci showed PTEN loss, and of these 66% (N=27/41) showed heterogeneous PTEN loss. In ERG homogeneously positive cases, any PTEN loss occurred in 56% (N=23/41) of cases, and of these 65% (N=15/23) showed heterogeneous loss. In ERG-negative tumors, 51.5% (N=17/33) showed PTEN loss, and of these 64.7% (N=11/17) showed heterogeneous PTEN loss. In a subset of cases, genomic deletions of PTEN were verified by fluorescence in situ hybridization in regions with PTEN protein loss as compared with regions with intact PTEN protein, which did not show PTEN genomic loss.
Conclusions:
These results support the concept that PTEN loss tends to occur as a subclonal event within a given established prostatic carcinoma clone after ERG gene fusion. The combination of ERG and PTEN IHC staining can be used as a simple test to ascertain PTEN and ERG gene rearrangement status within a given prostate cancer in either a research or clinical setting.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 4 print issues and online access
$259.00 per year
only $64.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Greaves M, Maley CC . Clonal evolution in cancer. Nature 2012; 481: 306–313.
Hanahan D, Weinberg RA . Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674.
Jan M, Majeti R . Clonal evolution of acute leukemia genomes. Oncogene 2012; 32: 135–140.
Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 2005; 310: 644–648.
Di Cristofano A, Pandolfi PP . The multiple roles of PTEN in tumor suppression. Cell 2000; 100: 387–390.
Sansal I, Sellers WR . The biology and clinical relevance of the PTEN tumor suppressor pathway. J Clin Oncol 2004; 22: 2954–2963.
Yoshimoto M, Ludkovski O, DeGrace D, Williams JL, Evans A, Sircar K et al. PTEN genomic deletions that characterize aggressive prostate cancer originate close to segmental duplications. Genes Chromosomes Cancer 2012; 51: 149–160.
Reid AH, Attard G, Brewer D, Miranda S, Riisnaes R, Clark J et al. Novel, gross chromosomal alterations involving PTEN cooperate with allelic loss in prostate cancer. Mod Pathol 2012; 25: 902–910.
Furusato B, Tan SH, Young D, Dobi A, Sun C, Mohamed AA et al. ERG oncoprotein expression in prostate cancer: clonal progression of ERG-positive tumor cells and potential for ERG-based stratification. Prostate Cancer Prostatic Dis 2010; 13: 228–237.
Mehra R, Han B, Tomlins SA, Wang L, Menon A, Wasco MJ et al. Heterogeneity of TMPRSS2 gene rearrangements in multifocal prostate adenocarcinoma: molecular evidence for an independent group of diseases. Cancer Res 2007; 67: 7991–7995.
Barry M, Perner S, Demichelis F, Rubin MA . TMPRSS2-ERG fusion heterogeneity in multifocal prostate cancer: clinical and biologic implications. Urology 2007; 70: 630–633.
Furusato B, Gao CL, Ravindranath L, Chen Y, Cullen J, McLeod DG et al. Mapping of TMPRSS2-ERG fusions in the context of multi-focal prostate cancer. Mod Pathol 2008; 21: 67–75.
Bismar TA, Yoshimoto M, Vollmer RT, Duan Q, Firszt M, Corcos J et al. PTEN genomic deletion is an early event associated with ERG gene rearrangements in prostate cancer. BJU Int 2011; 107: 477–485.
Carver BS, Tran J, Chen Z, Carracedo-Perez A, Alimonti A, Nardella C et al. ETS rearrangements and prostate cancer initiation. Nature 2009; 457: E1.
Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS et al. Tomlins et al. reply. Nature 2009; 457: 2.
Park K, Tomlins SA, Mudaliar KM, Chiu YL, Esgueva R, Mehra R et al. Antibody-based detection of ERG rearrangement-positive prostate cancer. Neoplasia 2010; 12: 590–598.
van Leenders GJ, Boormans JL, Vissers CJ, Hoogland AM, Bressers AA, Furusato B et al. Antibody EPR3864 is specific for ERG genomic fusions in prostate cancer: implications for pathological practice. Mod Pathol 2011; 24: 1128–1138.
Falzarano SM, Zhou M, Hernandez AV, Klein EA, Rubin MA, Magi-Galluzzi C . Single focus prostate cancer: pathological features and ERG fusion status. J Urol 2011; 185: 489–494.
Chaux A, Albadine R, Toubaji A, Hicks J, Meeker A, Platz EA et al. Immunohistochemistry for ERG expression as a surrogate for TMPRSS2-ERG fusion detection in prostatic adenocarcinomas. Am J Surg Pathol 2011; 35: 1014–1020.
Lotan TL, Gurel B, Sutcliffe S, Esopi D, Liu W, Xu J et al. PTEN protein loss by immunostaining: analytic validation and prognostic indicator for a high risk surgical cohort of prostate cancer patients. Clin Cancer Res 2011; 17: 6563–6573.
Sangale Z, Prass C, Carlson A, Tikishvili E, Degrado J, Lanchbury J et al. A robust immunohistochemical assay for detecting PTEN expression in human tumors. Appl Immunohistochem Mol Morphol 2011; 19: 173–183.
Lotan TL, Gupta NS, Wang W, Toubaji A, Haffner MC, Chaux A et al. ERG gene rearrangements are common in prostatic small cell carcinomas. Mod Pathol 2011; 24: 820–828.
Qu X, Randhawa G, Friedman C, O'Hara-Larrivee S, Kroeger K, Dumpit R et al. A novel four-color fluorescence in situ hybridization assay for the detection of TMPRSS2 and ERG rearrangements in prostate cancer. Cancer Genet 2013; 206: 1–11.
Yoshimoto M, Ding K, Sweet JM, Ludkovski O, Trottier G, Song KS et al. PTEN losses exhibit heterogeneity in multifocal prostatic adenocarcinoma and are associated with higher Gleason grade. Mod Pathol 2012; 26: 435–447.
Vogelstein B, Kinzler KW . The multistep nature of cancer. Trends Genet 1993; 9: 138–141.
Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 2012; 481: 506–510.
Snuderl M, Fazlollahi L, Le LP, Nitta M, Zhelyazkova BH, Davidson CJ et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell 2011; 20: 810–817.
Han B, Mehra R, Lonigro RJ, Wang L, Suleman K, Menon A et al. Fluorescence in situ hybridization study shows association of PTEN deletion with ERG rearrangement during prostate cancer progression. Mod Pathol 2009; 22: 1083–1093.
Yoshimoto M, Cunha IW, Coudry RA, Fonseca FP, Torres CH, Soares FA et al. FISH analysis of 107 prostate cancers shows that PTEN genomic deletion is associated with poor clinical outcome. Br J Cancer 2007; 97: 678–685.
Reid AH, Attard G, Ambroisine L, Fisher G, Kovacs G, Brewer D et al. Molecular characterisation of ERG, ETV1 and PTEN gene loci identifies patients at low and high risk of death from prostate cancer. Br J Cancer 2010; 102: 678–684.
Albadine R, Latour M, Toubaji A, Haffner M, Isaacs WB, AP E et al. TMPRSS2-ERG gene fusion status in minute (minimal) prostatic adenocarcinoma. Mod Pathol 2009; 22: 1415–1422.
Toubaji A, Albadine R, Meeker AK, Isaacs WB, Lotan T, Haffner MC et al. Increased gene copy number of ERG on chromosome 21 but not TMPRSS2-ERG fusion predicts outcome in prostatic adenocarcinomas. Mod Pathol 2011; 24: 1511–1520.
Liu W, Laitinen S, Khan S, Vihinen M, Kowalski J, Yu G et al. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat Med 2009; 15: 559–565.
Suzuki H, Freije D, Nusskern DR, Okami K, Cairns P, Sidransky D et al. Interfocal heterogeneity of PTEN/MMAC1 gene alterations in multiple metastatic prostate cancer tissues. Cancer Res 1998; 58: 204–209.
Perner S, Mosquera JM, Demichelis F, Hofer MD, Paris PL, Simko J et al. TMPRSS2-ERG fusion prostate cancer: an early molecular event associated with invasion. Am J Surg Pathol 2007; 31: 882–888.
Yoshimoto M, Cutz JC, Nuin PA, Joshua AM, Bayani J, Evans AJ et al. Interphase FISH analysis of PTEN in histologic sections shows genomic deletions in 68% of primary prostate cancer and 23% of high-grade prostatic intra-epithelial neoplasias. Cancer Genet Cytogenet 2006; 169: 128–137.
Lotan TL, Gumuskaya B, Rahimi H, Hicks JL, Iwata T, Robinson BD et al. Cytoplasmic PTEN protein loss distinguishes intraductal carcinoma of the prostate from high-grade prostatic intraepithelial neoplasia. Mod Pathol 2012; (e-pub ahead of print).
Acknowledgements
We thank the members of the Johns Hopkins Urological Specimen Repository and Database, and the Tissue Microarray Core at the Sidney Kimmel Comprehensive Cancer Center and Department of Pathology for help with tissue samples included. Funding was provided by the NCI Prostate SPORE P50CA58236, the DOD Prostate Bio specimen Repository Network, The Prostate Cancer Foundation and the Patrick C. Walsh Prostate Cancer Fund, of which AMD is the Peter J. Sharp Foundation Scholar, and a generous gift from David M. Koch.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
AMD is currently an employee of Predictive Biosciences. However, no funding or other support was provided by the company for any of the work in this manuscript. The terms of the relationship between AMD and Predictive Biosciences are managed by the Johns Hopkins University in accordance with its conflict-of-interest policies. All other authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Gumuskaya, B., Gurel, B., Fedor, H. et al. Assessing the order of critical alterations in prostate cancer development and progression by IHC: further evidence that PTEN loss occurs subsequent to ERG gene fusion. Prostate Cancer Prostatic Dis 16, 209–215 (2013). https://doi.org/10.1038/pcan.2013.8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/pcan.2013.8
Keywords
This article is cited by
-
Immunohistochemical expression of mismatch repair proteins (MSH2, MSH6, MLH1, and PMS2) in prostate cancer: correlation with grade groups (WHO 2016) and ERG and PTEN status
Virchows Archiv (2019)
-
Clinical implications of PTEN loss in prostate cancer
Nature Reviews Urology (2018)
-
SPINK1 Overexpression in Localized Prostate Cancer: a Rare Event Inversely Associated with ERG Expression and Exclusive of Homozygous PTEN Deletion
Pathology & Oncology Research (2017)
-
Heterogeneity of ERG expression in prostate cancer: a large section mapping study of entire prostatectomy specimens from 125 patients
BMC Cancer (2016)
-
Loss of PTEN expression in ERG-negative prostate cancer predicts secondary therapies and leads to shorter disease-specific survival time after radical prostatectomy
Modern Pathology (2016)