Abstract
Traf2- and Nck-interacting kinase (TNIK) is one of the germinal center kinase family members involved in cytoskeleton organization and neuronal dendrite extension. Emerging evidence supports that TNIK is essential for activation of WNT signaling pathway in colon cancer growth. To search for novel genetic aberrations that drive carcinogenesis, we performed microarray-based comparative hybridization assay for gene copy number variations in primary tumor samples. Our data showed that TNIK gene was amplified in 7% (8/106) of Chinese gastric cancer patients. Theses amplifications were confirmed by fluorescence in situ hybridization analysis. PAMC82 human gastric cancer and T47D human breast cancer cell lines with TNIK amplification were identified to further understand the function of TNIK gene amplification. RNA-interference-mediated silencing of TNIK resulted in significant inhibition of cell growth and induction of cell death in TNIK-amplified, but not in TNIK-non-amplified, cell lines tested. This selective sensitivity to the TNIK inhibition was also observed under the effect of a small-molecule TNIK inhibitor. Furthermore, our data indicated that TNIK’s role in gastric cancer growth was not dependent on Wnt signaling but rather was involved in AKT activation and cell autophagy. Together, our results suggest that TNIK is a novel therapeutic target in gastric cancer and TNIK amplification can be potentially used for patient selection.
Similar content being viewed by others
Introduction
TNIK is a member of germinal center kinases possessing an N-terminal kinase domain. It was first identified as a tumor necrosis factor receptor–associated factor-2 and Nck-interacting kinase, which can specifically activates the c-Jun N-terminal kinase (JNK) pathway when transfected into cells.1 In addition, TNIK can physically associate with Rap2 to regulate cytoskeleton organization and cell spreading.1, 2 By taking a proteomics approach, Mahmoudi et al.3 observed TNIK as a T-cell transcription factor 4 (TCF4) interactor in the proliferative crypts of mouse small intestine. In the Wnt-activated intestinal crypts and colorectal cancer cells, TNIK is localized in the nuclei and functions as transcription activator to promoters of Wnt target genes in a β-catenin-dependent manner. In vitro binding and kinase assays indicated that TNIK directly binds with both TCF4 and β-catenin and phosphorylates TCF4. Exogenous expression of TNIK kinase inactive mutants abrogates TCF/LEF (lymphoid enhancer-binding factor)-driven transcription, suggesting that its kianse activity is essential in TCF/LEF-driven transcriptional activation of Wnt signaling.
The compelling data reported by Shitashige et al.4 further documented TNIK’s importance in Wnt signaling and growth of colorectal cancer cells. In their study, the authors showed that the wild-type TNIK, but not its catalytically inactive mutant, phosphorylated the conserved serine 154 residue of TCF4. Silencing of TNIK expression by small interfering RNA (siRNA) suppressed proliferation of colorectal cancer cells in vitro and the tumor growth in vivo. The growth inhibition could be rescued by reintroduction of the catalytic domain of TNIK, confirming the requirement of its enzymatic activity in maintenance of colorectal cancer growth. Together, data from this investigation suggest that TNIK is a potential target for the development of a pharmacological kinase inhibitor for the treatment of colorectal cancer with aberrant Wnt signaling. However, the role of TNIK in tumorigenesis outside of colorectal cancer is still unknown.
Given the multiplex biological functions of TNIK besides regulating Wnt signaling, it is speculated that this kinase may also be involved in tumorigenesis of other cancer types. Indeed, by screening gene copy number variations, we found that TNIK is amplified in Chinese gastric tumors. Our results by using siRNA-mediated gene silencing and a small-molecule TNIK inhibitor also suggest that TNIK is a potential target in solid tumors beyond colorectal cancer.
Results
Amplification of TNIK gene in Chinese gastric cancer
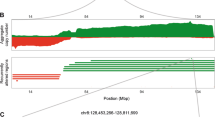
Amplification of oncogenes is one of the major genetic aberrations driving tumorigenesis. To identify potential anticancer targets with gene copy number gain, we performed array comparative genomic hybridization (aCGH) analysis in a total of 131 Chinese gastric cancer samples. We set up the criteria of gene amplification as log ratio ⩾0.8. TNIK was identified as one of the amplified genes in 4% (5/131) of gastric cancer samples (data not shown). Because aCGH assay cannot distinguish the exact location of the duplicated chromosomes, we further confirmed the TNIK amplification by performing fluorescence in situ hybridization (FISH) analysis on tumor microarray sections. In this study, we referred the Food and Drug Administration-approved EGFR FISH criteria for gene amplification, defined as TNIK/CEP3 ratio ⩾2 or the presence of clusters in ⩾10% tumor cells. Meanwhile immunohistochemistry (IHC) staining was performed to monitor the protein expression. Hence, the FISH-defined TNIK amplification was observed in 7% (8/107) Chinese gastric cancer, which was correlated with increased TNIK protein expression (Table 1; Figures 1a and b).
Representative images of TNIK FISH and IHC analysis of TNIK and β-catenin IHC analysis on TNIK-amplified and -non-amplified gastric cancer samples. (a) Images from TNIK FISH analysis. Green signals represent TNIK gene, red signals are CEP3 as internal control and blue are DAPI staining for nuclei. (b) IHC staining of TNIK. (c) IHC staining of β-catenin. C, M and N represent cytoplasm, cytoplasmic membrane and nucleus localization, respectively.
TNIK amplification is not associated with activation of Wnt signaling
Previous study indicated that TNIK is required for the activation of Wnt signaling in colorectal cancer.3 Thus, we assessed β-catenin nucleus localization, a hallmark of activation of Wnt signaling, in the TNIK-amplified tumors by IHC staining. However, the nucleus localization of β-catenin was only observed in one of the TNIK-amplified tumors tested (Table 1; Figure 1c). These results suggest that TNIK amplification is not absolutely associated with Wnt signaling in gastric cancer.
Identification of TNIK-amplified cell lines
We next screened cell lines with TNIK amplification by FISH analysis for further functional studies. Human gastric cancer cell line PAMC82 and breast cancer cell line T47D were identified with TNIK amplification (Table 2 and Figure 2a). Two TNIK-non-amplified gastric cancer cell lines SNU638 and AZ521 were chosen as controls. The expression levels of TNIK protein in these cell lines and β-catenin localization were measured by western blotting and IHC analysis, respectively. As shown in Figure 2b, significantly higher levels of TNIK protein were detected in both PAMC82 and T47D cells compared with that in the control cells (the relative expression levels were compared in Supplementary Figure 1). Consistent with the observation from TNIK-amplified gastric tumors, the nucleus localization of β-catenin was observed in the TNIK-non-amplified AZ521, but not in the TNIK-amplified PAMC-82 and T47D cells (Figure 2c), suggesting a TNIK-independent mechanism for Wnt activation in gastric cancer cells.
Profiling of PAMC82, T47D, SNU638 and AZ521 cell lines. (a) The amplification of TNIK in the three cell lines were measured by FISH analysis as described above. (b) TNIK protein expression was detected by western blotting. Cell lysates collected from PAMC82, T47D, SNU638 and AZ521 cells were subjected to western blotting analysis with the indicated antibodies. (c) The expression and localization of β-catenin were detected by IHC staining on the four cell lines as described above. *Amp means amplification. The data presented here are representative from three independent experiments.
siRNA-mediated gene silencing led to reduced cell growth and increased cell death in TNIK-amplified but not in the TNIK-non-amplified tumor cells
To test whether TNIK amplification drives cell proliferation/survival, siRNA-mediated TNIK silencing was applied. TNIK-amplified PAMC82 and T47D cells and TNIK-non-amplified SNU638 and AZ521 cells were transiently transfected with the siRNA directly against TNIK, and cell proliferation/survival was monitored by both Acumen and IncuCyte photography. As shown in Figure 3, introduction of TNIK-specific siRNAs, but not the scrambled control siRNAs, resulted in significant reduction of cell growth and induction of cell death. Consistently, the western blotting analysis conducted in parallel demonstrated the significant reduction of TNIK protein expression, correlating with the cell growth inhibition.
Knockdown of TNIK by siRNA led to cell growth inhibition and death. (a, b) siRNAs against TNIK and scramble sequence were introduced in cells by transfection with HiPerFect. Cell proliferation (a) and death (b) were monitored by Acumen analysis after transfection for 96 h. (c) Knockdown of TNIK protein expression by TNIK siRNAs. Cells were transfected with different TNIK siRNAs or scrambled siRNA for 48 h, and cell lysates were collected for western blotting analysis using anti-TNIK antibody. The data represented here are representative from three independent experiments. UN, untreated cells, NC, scrambled control, S2, TNIKsiRNA 2, S3, TNIKsiRNA3. *P<0.05, **P<0.01.
TNIK amplification is involved in phosphatidylinositol 3′-kinase (PI3K)/AKT pathway and regulation of cell autophagy
Tesshi et al. recently reported the development of a potent and selective TNIK small-molecule kinase inhibitor.5 We wanted to test the sensitivity of TNIK-amplified and -non-amplified cells to this TNIK inhibitor. Cells seeded in 96-well plates were treated with TNIK inhibitor at different concentrations for 72 h, and cell viability was measured by the Hoechst and propidium iodide (PI) staining assay. As shown in Figure 4, PAMC82 and T47D cells harboring TNIK amplification were sensitive to TNIK inhibitor with IC50 of 1.77 and 0.385 μmol/l, respectively. In contrast, TNIK-non-amplified AZ-521 cells were insensitive to TNIK inhibition with IC50 >30 μmol/l. However, the TNIK-non-amplified SNU638 cells were also sensitive to TNIK inhibitor with IC50 of 0.9 μmol/l, suggesting an off-target effect from the TNIK inhibitor in this cell type. We further investigated the potential mechanism for the growth inhibition. PAMC82 and AZ521 cells were treated with TNIK inhibitor at different concentrations for 2 h, and cell lysates were subjected to western blotting analysis. In consistent with the growth-inhibitory effect, treatment with TNIK inhibitor from 1 to 10 μmol/l led to a significant downregulation of phosphorylation of AKT in TNIK-amplified PAMC82, but not in the TNIK-non-amplified AZ521 cells, suggesting a possible mechanism of TNIK amplification in cell growth. Meanwhile, we also examined the levels of poly ADP-ribose polymerase (PARP), cleaved caspase 3 and LC3, the hallmarks of cell apoptosis and autophagy, respectively. Interestingly, the treatment of TNIK inhibitor resulted in upregulation of LC3 (Figure 5), but had no effect on PARP and caspase 3 (data not shown), suggesting a protective role of TNIK in cell autophagy. Although the differential effects observed by using TNIK inhibitor support a direct role of TNIK on AKT activation and LC3 induction, it is still suspicious that this effect may be due to the off-target effect of the small-molecule kinase inhibitor. Thus, we further tested the effects by siRNA-mediated TNIK knockdown in PAMC82 and AZ521 cells. As shown in Figure 5b, downregulation of TNIK resulted in reduced pAKT and induction of LC3 in PAMC82, but not in the TNIK-non-amplified AZ521 cells, confirming the role of TNIK in activation of AKT pathway and protection of cell autophagy. Interestingly, the basal level of phosphor AKT in the TNIK-amplified PAMC82 and T47D cells were 2.4- and 6-folders higher than that in the SNU638 and AZ521 cells, respectively (Supplementary Figure 1), supporting a role of TNIK in the regulation of AKT activation.
Suppression of AKT phosphorylation and induction of LC3 by TNIK inhibitor or siRNA-mediated TNIK knockdown. (a) PAMC82 and AZ521 cells were treated with TNIK inhibitor at the indicated concentrations for 2 h, and cell lysates were collected for western blotting analysis with the indicated antibodies. (b) PAMC82 and AZ521 cells were transfected with siRNA against TNIK or scrambled, and cell lysates were collected for western blotting analysis with the indicated antibodies. The data presented here are representative from three independent experiments. UN, untreated cells, NC, scrambled control, S2, TNIKsiRNA 2, S3, TNIKsiRNA3.
Profiling of genetic aberrations by whole-transcriptome sequencing
To further uncover any genetic aberrations that might confer the oncogenic role in TNIK-amplified gastric cancer, we performed whole-transcriptome sequencing (RNA sequencing) of 6 available gastric cancer samples with TNIK amplification, 13 TNIK-non-amplified gastric cancer samples and 6 adjacent normal gastric tissues using the Illumina HiSeq2000 system (Illumina, San Diego, CA, USA). All the specimens were surgically removed from Chinese gastric cancer patients who were treatment naive. We first searched for the known genetic-driving aberrations reported in the six TNIK-amplified samples. As shown in Table 3, the mutations of CTNNB1 (M662I) and adenomatous polyposis coli (APC) gene (p.R216*) were found in GC-274 primary tumor (1/6), while HER2 amplification and FGFR2 amplification were observed in GC-268 and GC-288 tumors, respectively, indicating the coexistence of TNIK amplification with other oncogenic drivers in some of the cases (3/6). These results further support the Wnt-independent function of TNIK in gastric cancer.
To explore additional pathways affected by TNIK amplification, we compared the gene expression profiles between the 6 gastric tumors with TNIK amplification and the 13 tumors without TNIK non-amplification as well as the 6 normal tissues. As shown in Table 4, by using a 10-folders cutoff, the upregulation of a total of 46 genes was observed (P<0.05), whereas only two downregulated genes (COMP and DERL3) were found. Interestingly, PI3KCA was found as one of the upregulated genes, suggesting a potential mechanism for the increased activation of AKT mediated by TNIK.
Discussion
The current development of targeted cancer therapy is featured by companion diagnosis based on specific biomarkers for patient selection. The design of such a personalized health-care strategy is greatly facilitated and accelerated by the broad application of genome-wide genetic profiling technologies for identification of cancer-driver genes and biomarkers. By using the aCGH approach to search for new genetic aberrations in gastric cancer, we demonstrated for the first time the amplification of TNIK, a protein kinase involved in Wnt signaling in intestinal crypts and colorectal cancer cells. However, the amplification of TNIK is not associated with the activation of Wnt signaling, defined by β-catenin nucleus localization, in 89% (8/9) of TNIK-amplified gastric tumors tested and lack of gene mutations for activation of Wnt signaling in 83% (5/6) of the tested TNIK-amplified gastric tumors. Silencing of TNIK by RNA interference led to growth inhibition and cell death in TNIK-amplified, but not in the TNIK-non-amplified, cell lines tested (Figure 3). This observation was further confirmed by using a small-molecule TNIK kinase inhibitor, supporting an essential role of TNIK kinase activity for tumorigenesis in gastric cancer carrying TNIK amplification.
Wnt signaling pathways are highly conserved and are critical in regulating a number of physiological functions, including cell proliferation, differentiation, motility, survival and/or apoptosis.6, 7, 8 These pathways can be divided into the well-characterized canonical (Wnt/β-catenin pathway) and the noncanonical pathways (the planar cell polarity pathway, the Wnt/Ca2+ pathway, the protein kinase A pathway). The activity of catenin signaling pathway relies on the cytoplasmic levels of β-catenin, which is regulated by ubiquitin-proteasome-mediated degradation involved in a multiprotein ‘destruction’ complex containing axin, APC and glycogen synthase kinase-3β. Normally, Wnt signaling is quiescent due to a low level of cytoplasmic β-catenin. Upon binding of Wnt proteins to its receptor complex, a protein downstream of the receptor complex is phosphorylated, which first leads to inhibition of glycogen synthase kinase-3β, followed by the accumulation of nonphosphorylated β-catenin in the cytoplasm and eventual translocation of β-catenin into the nucleus. In the nucleus, β-catenin forms a complex with members of the TCF/LEF family of transcription factors to control gene expression.9
Previous studies indicated that TNIK is an essential component in Wnt signaling to control growth and a potential therapeutic target for colorectal cancer.3, 4 However, a few lines of evidences from our study support a Wnt-independent oncogenic role of TNIK amplification in gastric cancer. First, in several tested gastric tumors and cell lines (Tables 1 and 3 and Figures 1 and 2), the amplification of TNIK is not associated with β-catenin nucleus localization and mutations of genes involving in activation of Wnt signaling (CTNNB1, AXIN1 and APC). Second, the TNIK-non-amplified AZ521 gastric cancer cell line with active Wnt signaling is insensitive to TNIK-specific siRNA and small-molecule inhibitor. Consistently, we could not detect significant transcriptional changes of genes controlled by Wnt signaling in the TNIK-amplified cells after shRNA-mediated TNIK knockdown (data not shown). In fact, results reported by other groups also support more broad roles of TNIK in other organs and tissues. For instance, in neurons, TNIK was found to be involved in the regulation of neurite growth and neuronal structure.10, 11 Recently, Shkoda et al.12 reported that TNIK physically associates with the latent membrane protein 1 signalosome in primary human B-cells infected with the Epstein–Barr tumor virus (EBV) to control EBV-mediated B-cell proliferation, survival and transformation. This regulation is involved in TRAF6 (tumor necrosis factor receptor-associated factor 6)-dependent nuclear factor-κB and JNK pathways. However, we could not observe significant impact of TNIK inhibition on nuclear factor-κB and JNK pathways in TNIK-amplified cells (data not shown), suggesting the selective involvement of the amplified TNIK in different pathways.
Although RNA interference provides us with a very specific and an efficient tool for the evaluation of gene functions in many instances, it still possesses some limitations compared with small-molecule inhibitors for the assessment of the biological effects of a particular enzyme in a quantitative and timely manner. Therefore, we utilized a small-molecule kinase inhibitor of TNIK, a novel aminothiazole derivative, which is potently against TNIK in in vitro enzymatic assay with IC50 of 9 nmol/l and a reasonable selectivity over 30-folds against the majority of a panel of 50 kinases profiled. In this study, we found that the two TNIK-amplified cell lines PAMC82 and T47D, but not the TNIK-non-amplified AZ521, are very sensitive to this inhibitor, consistent with the effect of shRNA-mediated TNIK protein knockdown. It is noteworthy that the treatment of TNIK-amplified PAMC82, but not AZ521, cells with the TNIK inhibitor led to suppression of pAKT and induction of LC3, the hallmark of cell autophagy, indicating a TNIK-dependent effect. Autophagy is a catabolic process involving the degradation of a cell’s own components through the lysosomal machinery and can be induced by suppression of PI3K/Akt/mTOR pathway.13, 14 Because the TNIK inhibitor used in this study also has inhibitory activity against other kinases such as kinase insert domain receptor (KDR), the effect of TNIK inhibition on pAKT and LC3 might be due to its off-target effect. To rule out of this possibility, we further demonstrated the similar effects of TNIK on pAKT and LC3 by using siRNA-mediated TNIK knockdown (Figure 5b). Although the expression of KDR protein was not detectable by western blotting in the cell lines used in this study (data not shown), we could not completely rule out the off-target effects on suppression of cell proliferation by the TNIK inhibitor. In fact, the TNIK-non-amplified SNU638 cells are sensitive to the TNIK inhibitor, which must be due to other mechanisms (Figure 4). It is also noteworthy that the inhibition of pAKT alone by TNIK inhibition may not be sufficient for cell growth suppression, because both PAMC82 and AZ521 cells were not sensitive to AKT inhibitor AZD5363 in vitro.15 This result suggests that other un-identified TNIK downstream pathway(s) must also contribute to the biological function of TNIK in gastric cancer. At this point, the mechanism for pAKT inhibition by TNIK inhibitor is not fully understood. Our RNAseq analysis indicated the upregulation of PI3KCA mRNA in TNIK-amplified tumors compared with that in the TNIK-non-amplified gastric tumors and the adjacent normal tissues, suggesting a potential mechanism for TNIK-mediated activation of AKT pathway. Further studies for understanding the regulation may bring us new insights for Wnt-independent functions of TNIK. Recently, Ho et al.16 developed a series of potent and selective TNIK inhibitors based on a 4-phenyl-2-phenylaminopyridine scaffold. By using these inhibitors, the authors indicated that pharmacological inhibition of TNIK kinase activity has minimal effects on either Wnt signaling-mediated transcription or viability of the tested colorectal cancer cells. These data together with our observations support the existence of a TNIK-independent activation of Wnt signaling.
Materials and methods
Cell culture and antibodies
Human gastric cancer cell lines PAMC82 and AZ521 were obtained from Beijing Tumor Hospital (Beijing, China) and KCLB (Korean Cell Line Bank, Seoul, Korea), respectively. Human breast cancer cell line T47D was obtained from ATCC (American Type Culture Collection, Rochville, MD, USA). SNU-638 cells were obtained from Korean Cell Line Bank. T47D cell line was cultured in Dulbecco’s Modified Eagle’s Medium (Invitrogen, Carlsbad, CA, USA), supplemented with 10% fetal bovine serum (Invitrogen), 2 mmol/l L-Glutamine (L-Glu, Fisher Scientific, Ottama, ON, Canada). PAMC82, SNU638 and AZ521 cells were maintained in RPMI 1640 (Invitrogen) supplemented with 10% fetal bovine serum and 2 mmol/l L-Glutamine. All cells were maintained in a humidified incubator with 5% CO2 at 37 °C. Antibodies used in this study were obtained from the following sources: TNIK (GeneTex, Irvine, CA, USA, Cat. GTX13141), phospho-Akt (S473) (Dako, Produktionsvej, Glostrup, Denmark Cat.M3628), phospho-Erk1/2 (Thr202/Tyr204) (Cell Signaling Technologies, Danvers, MA, USA, Cat. 4370), AKT (Cell Signaling Technologies, Cat. 9272), LC3 (Novus Biologics, Littleton, CO, USA, Cat.NB100-2220), and GAPDH (glyceraldehyde 3-phosphate dehydrogenase; Cell Signaling Technologies, Cat. 2118).
Compound synthesis
TNIK inhibitor was synthesized by PharmaResources (Shanghai, China) according to the patent WO 2010/064111 A1.5 The structure was disclosed in Supplementary Figure 2. The molecular weight was determined by liquid chromatography–mass spectrometry, and the purity was measured by high-performance liquid chromatography. The purity of TNIK inhibitor used for the in vitro cellular assay was >99%.
Patients and tissue specimens
Chinese gastric cancer specimens were collected at surgery from gastric cancer patients with postoperative pathological confirmation in Peking University Cancer Hospital (2007∼2010), Shanghai Renji Hospital, Shanghai, China. Written informed consent was provided by each patient, and the study was approved by the ethical committee of the Hospital. Harvested fresh gastric cancer specimens were fixed in 10% buffered formalin within 30 min and embedded into paraffin (FFPE). In all, 2∼4 × 0.6 mm2 cores from each tumor and two from matched adjacent mucosa were randomly selected for tissue microarray construction.
siRNA, transfection and proliferations assays
Cells were seeded at a density of 2500 cells/well on 96-well plates. Predesigned siRNA for human TNIK (5′-GCGCAAACAATTGGAAGAA-3′, 5′-GATCTGACGGCATTAGCCA-3′, 5′-CTAAGGATGTGGTGCTCCA-3′) and the Allstar negative control siRNA (5′-AACACAGTGGAGCGAATTCCT-3′) were purchased from Qiagen (Valencia, CA, USA). Cells were transfected with 10 nmol/l siRNA, using 1: 1 HiPerFect Transfection Reagent (Qiagen) according to the manufacturer’s guidelines. Cell viability was measured by the Hoechst and PI staining assay. Briefly, cells were incubated with 10 μmol/l of Hoechest 34580 (Invitrogen) and 1.5 μmol/l of PI for 30 min and then detected by Acumen X3 (TTP, Melbourm, Hertfordshire, UK). Viabilities were expressed as a percentage relative to the untreated controls. Statistical significance was evaluated using a one-tailed, two-sample t-test. P<0.05 was considered statistically significant.
Western blotting analysis
Total cellular extracts from cell lines were prepared in sodium dodecyl sulfate lysis buffer supplemented with protease inhibitors and phosphotase inhibitors (Sigma, St Louis, MO, USA). Protein samples were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and blotted onto polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA). After incubation with indicated antibodies at 4 °C overnight, the blots were detected with the relevant horseradish peroxidase-conjugated anti-mouse or anti-rabbit immunoglobulin G antibody and enhanced chemiluminescence (GE Healthcare, Piscataway, NJ, USA).
aCGH microarray analysis
DNA was extracted using the QIAamp DNA Mini Kit (Qiagen) according to the manufacturer’s instructions. aCGH analysis was performed using the Agilent Human Genome 244K Microarray Chip (Agilent Technologies, Folsom, CA, USA), with pooled human genomic DNA (Promega, Madison, WI, USA) as reference. Labeling, hybridization, washes and data analysis were performed according to the protocol provided by Agilent (Protocol version 4.0, June 2006). Graphical overviews were obtained using the CGH Analytics software Nexus DNA copy number version 3.0 (El Sequndo, CA, USA).
FISH
FISH analysis performed on 4-μm tissue microarray section was used to assess the presence of gene amplification. The TNIK FISH probe was generated in house by directly labeling a mixture of BAC RP11-466C5, RP11-26K1 and RP11-933C17 (Invitrogen) DNA with Spectrum Green (ENZO, 02N32-050, Farmingdale, NY, USA) using a nick translation-based method (Abbott, Des Plaines, IL, USA, 07J00-001) according to manufacturer’s instructions. Pericentromeric Spectrum Orange-labeled chromosome 3 probe (CEP3, Vysis, Downess Grove, IL, USA, 32-110003) was used as an internal control. FISH was performed as described previously.17 Sections were deparaffinized and pretreated using the SpotLight Tissue Kit (Invitrogen, 00-8401) according to manufacturer’s instructions. Sections and TNIK/CEP3 probes were co-denaturated at 80 °C for 5 min and hybridized at 37 °C for 48 h. The excess probe was removed with post-hybridization wash buffer (0.3% NP40/1 × SSC) at 75.5 °C for 5 min and then with 2 × SSC at room temperature for 2 min. Sections were counterstained with 0.3 μg/ml DAPI (4,6-diamidino-2-phenylindole, Vector Lab., Burlingame, CA, USA, H-1200) and coverslipped. TNIK and CEP3 signals were scored under a fluorescence microscope (Olympus, BX61, Tokyo, Japan). TNIK gene amplification was defined as the presence of clusters in ⩾10% tumor cells or TNIK/CEP3 ratio ⩾2.
IHC
All tissues were collected following the standard procedure for FFPE blocks. β-Catenin IHC study was done on 3-μm xenograft sections and cell blocks and 4-μm human sections using a Lab Vision autostainer (Fremont, CA, USA). The procedures are described as follows: paraffin sections were dewaxed and rehydrated on a Leica XL autostainer (Heidelberger, Germany). Then the slides were treated antigen retrieval solution, pH9 (DAKO S2367) for 5 min followed by washing in running tap water for 5 min. Then the sections were rinsed in TBST (a mixture of Tris-buffered saline and Tween 20) and put on the LabVision autostainer. After incubation with endogenous peroxidase block (DAKO S2023) for 10 min, the slides were washed in TBST twice. The sections were then incubated with primary antibody (anti-β-catenin antibody, Epitomics 1247-1, Burlingame, CA, USA, 1:500 or anti-TNIK antibody, Sigma HPA012128, 1:200) for 60 min at room temperature and washed in TBST twice. After reaction with EnVision system-horseradish peroxidase-labeled polymer anti-rabbit (DAKO K4003) for 30 min and washing in TBST twice, the sections were developed in diaminobenzidine substrate (DAKO K3468) for 5 min and rinsed in tap water. Then the sections were counter stained, dehydrated, cleared and mounted with coverslips in Leica XL autostainer workstation. Scoring was established as follows: 0, if absence of staining was observed; 1+, if the tumor cells had weak staining; 2+, if tumor cells had moderate staining; and 3+, if tumor cells had strong staining. Tumors with expression scores of 2+ and 3+ were interpreted as positive and tumors with expression scores of 0 and 1+ were interpreted as negative. Given the heterogeneity of protein expression in tumor cells, the highest score from either of specimen cores was counted as the final result.
RNA sequencing
Total RNA quality and concentration was measured using an RNA Pico chip on a Bioanalyzer 2100 (Agilent). Normalized starting quantities of total RNA were then used to prepare Illumina sequencing libraries with the TruSeq RNA sample preparation kit (Illumina). Library preparation was performed according to the manufacturer’s instructions. The cDNA libraries were placed on an Illumina c-Bot for paired-end (PE) cluster generation according to the protocol outlined in the Illumina HiSeq Analysis User Guide. The template cDNA libraries (1.5 μg) were hybridized to a flow cell, amplified, linearized and denatured to create a flow cell with ssDNA ready for sequencing. Each flow cell was sequenced on an Illumina HiSeq Genome Analyzer. After 100-cycle PE sequencing run, bases and quality values were generated for each read with the current Illumina pipeline. Gene expression data (FPKM value) for each gene were calculated by the commercial software OmicSoft (Cary, NC, USA) (http://www.omicsoft.com/).
Change history
17 March 2014
This article has been corrected since Online Publication and an erratum has also been published.
References
Fu CA, Shen M, Huang BC, Lasaga J, Payan DG, Luo Y . TNIK, a novel member of the germinal center kinase family that activates the c-Jun N-terminal kinase pathway and regulates the cytoskeleton. J Biol Chem 1999; 274: 30729–30737.
Taira K, Umikawa M, Takei K, Myagmar BE, Shinzato M, Machida N et al. The Traf2- and Nck-interacting kinase as a putative effector of Rap2 to regulate actin cytoskeleton. J Biol Chem 2004; 279: 49488–49496.
Mahmoudi T, Li VS, Ng SS, Taouatas N, Vries RG, Mohammed S et al. The kinase TNIK is an essential activator of Wnt target genes. EMBO J 2009; 28: 3329–3340.
Shitashige M, Satow R, Jigami T, Aoki K, Honda K, Shibata T et al. Traf2- and Nck-interacting kinase is essential for Wnt signaling and colorectal cancer growth. Cancer Res 2010; 70: 5024–5033.
Yamada T, Shitashige M, Yokota K, Sawa M, Mariyama H TNIK inhibitor and the use. W02010/064111A1.
Nusse R, Varmus H . Three decades of Wnts: a personal perspective on how a scientific field developed. EMBO J 2012; 31: 2670–2684.
Pecina-Slaus N . Wnt signal transduction pathway and apoptosis: a review. Cancer Cell Int 2010; 10: 22.
Saito-Diaz K, Chen TW, Wang X, Thorne CA, Wallace HA, Page-McCaw A et al. The way Wnt works: components and mechanism. Growth Factors 2013; 31: 1–31.
Anastas JN, Moon RT . WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer 2013; 13: 11–26.
Hussain NK, Hsin H, Huganir RL, Sheng M . MINK and TNIK differentially act on Rap2-mediated signal transduction to regulate neuronal structure and AMPA receptor function. J Neurosci 2010; 30: 14786–14794.
Kawabe H, Neeb A, Dimova K, Young SM Jr, Takeda M, Katsurabayashi S et al. Regulation of Rap2A by the ubiquitin ligase Nedd4-1 controls neurite development. Neuron 2010; 65: 358–372.
Shkoda A, Town JA, Griese J, Romio M, Sarioglu H, Knofel T et al. The germinal center kinase TNIK is required for canonical NF-kappaB and JNK signaling in B-cells by the EBV oncoprotein LMP1 and the CD40 receptor. PLoS Biol 2012; 10: e1001376.
Maiese K, Chong ZZ, Shang YC, Wang S . Targeting disease through novel pathways of apoptosis and autophagy. Expert Opin Ther Targets 2012; 16: 1203–1214.
Parkhitko AA, Favorova OO, Henske EP . Autophagy: mechanisms, regulation, and its role in tumorigenesis. Biochemistry (Mosc) 2013; 78: 355–367.
Li J, Davies BR, Han S, Zhou M, Bai Y, Zhang J et al. The AKT inhibitor AZD5363 is selectively active in PI3KCA mutant gastric cancer, and sensitizes a patient-derived gastric cancer xenograft model with PTEN loss to Taxotere. J Transl Med 2013; 11: 241.
Ho KK, Parnell KM, Yuan Y, Xu Y, Kultgen SG, Hamblin S et al. Discovery of 4-phenyl-2-phenylaminopyridine based TNIK inhibitors. Bioorg Med Chem Lett 2013; 23: 569–573.
Xie L, Su X, Zhang L, Yin X, Tang L, Zhang X et al. FGFR2 gene amplification in gastric cancer predicts sensitivity to the selective FGFR inhibitor AZD4547. Clin Cancer Res 2013; 19: 2572–2583.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogenesis website .
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Yu, DH., Zhang, X., Wang, H. et al. The essential role of TNIK gene amplification in gastric cancer growth. Oncogenesis 3, e89 (2014). https://doi.org/10.1038/oncsis.2014.2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/oncsis.2014.2
Keywords
This article is cited by
-
Pharmacological blockage of transforming growth factor-β signalling by a Traf2- and Nck-interacting kinase inhibitor, NCB-0846
British Journal of Cancer (2021)
-
Protective autophagy promotes the resistance of HER2-positive breast cancer cells to lapatinib
Tumor Biology (2016)
-
Prognostic significance of Traf2- and Nck- interacting kinase (TNIK) in colorectal cancer
BMC Cancer (2015)