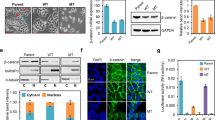
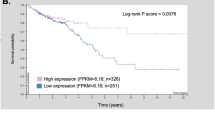
Abstract
E-cadherin and β-catenin are key proteins that are essential in the formation of the epithelial cell layer in the colon but their regulatory pathways that are disrupted in cancer metastasis are not completely understood. Mutated in colorectal cancer (MCC) is a tumour suppressor gene that is silenced by promoter methylation in colorectal cancer and particularly in patients with increased lymph node metastasis. Here, we show that MCC methylation is found in 45% of colon and 24% of rectal cancers and is associated with proximal colon, poorly differentiated, circumferential and mucinous tumours as well as increasing T stage and larger tumour size. Knockdown of MCC in HCT116 colon cancer cells caused a reduction in E-cadherin protein level, which is a hallmark of epithelial–mesenchymal transition in cancer, and consequently diminished the E-cadherin/β-catenin complex. MCC knockdown disrupted cell–cell adhesive strength and integrity in the dispase and transepithelial electrical resistance assays, enhanced hepatocyte growth factor-induced cell scatter and increased tumour cell invasiveness in an organotypic assay. The Src/Abl inhibitor dasatinib, a candidate anti-invasive drug, abrogated the invasive properties induced by MCC deficiency. Mechanistically, we establish that MCC interacts with the E-cadherin/β-catenin complex. These data provide a significant advance in the current understanding of cell–cell adhesion in colon cancer cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Pangon L, Sigglekow ND, Larance M, Al-Sohaily S, Mladenova DN, Selinger CI et al. The ‘mutated in colorectal cancer’ protein is a novel target of the UV-induced DNA damage checkpoint. Genes Cancer 2010; 1: 917–926.
Gamazon ER, Lamba JK, Pounds S, Stark AL, Wheeler HE, Cao X et al. Comprehensive genetic analysis of cytarabine sensitivity in a cell-based model identifies polymorphisms associated with outcome in AML patients. Blood 2013; 121: 4366–4376.
Matsumine A, Senda T, Baeg GH, Roy BC, Nakamura Y, Noda M et al. MCC, a cytoplasmic protein that blocks cell cycle progression from the G0/G1 to S phase. J Biol Chem 1996; 271: 10341–10346.
Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012; 487: 330–337.
Nishisho I, Nakamura Y, Miyoshi Y, Miki Y, Ando H, Horii A et al. Mutations of chromosome 5q21 genes in FAP and colorectal cancer patients. Science 1991; 253: 665–669.
Kohonen-Corish MR, Sigglekow ND, Susanto J, Chapuis PH, Bokey EL, Dent OF et al. Promoter methylation of the mutated in colorectal cancer gene is a frequent early event in colorectal cancer. Oncogene 2007; 26: 4435–4441.
Sigglekow ND, Pangon L, Brummer T, Molloy M, Hawkins NJ, Ward RL et al. Mutated in colorectal cancer protein modulates the NFkappaB pathway. Anticancer Res 2012; 32: 73–79.
Fukuyama R, Niculaita R, Ng KP, Obusez E, Sanchez J, Kalady M et al. Mutated in colorectal cancer, a putative tumor suppressor for serrated colorectal cancer, selectively represses beta-catenin-dependent transcription. Oncogene 2008; 27: 6044–6055.
Fu X, Li L, Peng Y . Wnt signalling pathway in the serrated neoplastic pathway of the colorectum: possible roles and epigenetic regulatory mechanisms. J Clin Pathol 2012; 65: 675–679.
Shukla R, Upton KR, Munoz-Lopez M, Gerhardt DJ, Fisher ME, Nguyen T et al. Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell 2013; 153: 101–111.
Lim L, Balakrishnan A, Huskey N, Jones KD, Jodari M, Ng R et al. MicroRNA-494 within an oncogenic microRNA megacluster regulates G1/S transition in liver tumorigenesis through suppression of mutated in colorectal cancer. Hepatology 2014; 59: 202–215.
Poursoltan P, Currey N, Pangon L, van Kralingen C, Selinger CI, Mahar A et al. Loss of heterozygosity of the Mutated in Colorectal Cancer gene is not associated with promoter methylation in non-small cell lung cancer. Lung Cancer 2012; 77: 272–276.
Starr TK, Allaei R, Silverstein KA, Staggs RA, Sarver AL, Bergemann TL et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science 2009; 323: 1747–1750.
Bard-Chapeau EA, Nguyen AT, Rust AG, Sayadi A, Lee P, Chua BQ et al. Transposon mutagenesis identifies genes driving hepatocellular carcinoma in a chronic hepatitis B mouse model. Nat Genet 2014; 46: 24–32.
Thiery JP, Acloque H, Huang RY, Nieto MA . Epithelial-mesenchymal transitions in development and disease. Cell 2009; 139: 871–890.
Shook D, Keller R . Mechanisms, mechanics and function of epithelial–mesenchymal transitions in early development. Mech Dev 2003; 120: 1351–1383.
Kalluri R, Neilson EG . Epithelial-mesenchymal transition and its implications for fibrosis. J Clin Invest 2003; 112: 1776–1784.
Lamouille S, Xu J, Derynck R . Molecular mechanisms of epithelial–mesenchymal transition. Nat Rev Mol Cell Biol 2014; 15: 178.
Behrens J, Birchmeier W, Goodman SL, Imhof BA . Dissociation of Madin-Darby canine kidney epithelial cells by the monoclonal antibody anti-arc-1: mechanistic aspects and identification of the antigen as a component related to uvomorulin. J Cell Biol 1985; 101: 1307–1315.
Nagafuchi A, Takeichi M . Cell binding function of E-cadherin is regulated by the cytoplasmic domain. EMBO J 1988; 7: 3679.
Ozawa M, Baribault H, Kemler R . The cytoplasmic domain of the cell adhesion molecule uvomorulin associates with three independent proteins structurally related in different species. EMBO J 1989; 8: 1711–1717.
Pangon L, Mladenova D, Watkins L, Van Kralingen C, Currey N, Al-Sohaily S et al. MCC inhibits beta-catenin transcriptional activity by sequestering DBC1 in the cytoplasm. Int J Cancer 2015; 136: 55–64.
Huang RY, Guilford P, Thiery JP . Early events in cell adhesion and polarity during epithelial-mesenchymal transition. J Cell Sci 2012; 125: 4417–4422.
Arnaud C, Sebbagh M, Nola S, Audebert S, Bidaut G, Hermant A et al. MCC, a new interacting protein for Scrib, is required for cell migration in epithelial cells. FEBS Lett 2009; 583: 2326–2332.
Pangon L, Van Kralingen C, Abas M, Daly RJ, Musgrove EA, Kohonen-Corish MR . The PDZ-binding motif of MCC is phosphorylated at position -1 and controls lamellipodia formation in colon epithelial cells. Biochim Biophys Acta 2012; 1823: 1058–1067.
Kim E, Niethammer M, Rothschild A, Jan YN, Sheng M . Clustering of Shaker-type K+ channels by interaction with a family of membrane-associated guanylate kinases. Nature 1995; 378: 85–88.
Tejedor FJ, Bokhari A, Rogero O, Gorczyca M, Zhang J, Kim E et al. Essential role for dlg in synaptic clustering of Shaker K+ channels in vivo. J Neurosci 1997; 17: 152–159.
Fleming M, Ravula S, Tatishchev SF, Wang HL . Colorectal carcinoma: pathologic aspects. J Gastrointest Oncol 2012; 3: 153–173.
Compton CC . Colorectal carcinoma: diagnostic, prognostic, and molecular features. Mod Pathol 2003; 16: 376–388.
Griffin MJ . Synchronization of some human cell strains by serum and calcium starvation. In Vitro 1976; 12: 393–398.
Calautti E, Cabodi S, Stein PL, Hatzfeld M, Kedersha N, Dotto GP . Tyrosine phosphorylation and src family kinases control keratinocyte cell–cell adhesion. J Cell Biol 1998; 141: 1449–1465.
von Bonsdorff C-H, Fuller SD, Simons K . Apical and basolateral endocytosis in Madin-Darby canine kidney (MDCK) cells grown on nitrocellulose filters. EMBO J 1985; 4: 2781.
Spina A, De Pasquale V, Cerulo G, Cocchiaro P, Della Morte R, Avallone L et al. HGF/c-MET axis in tumor microenvironment and metastasis formation. Biomedicines 2015; 3: 71–88.
Serrels A, Timpson P, Canel M, Schwarz JP, Carragher NO, Frame MC et al. Real-time study of E-cadherin and membrane dynamics in living animals: implications for disease modeling and drug development. Cancer Res 2009; 69: 2714–2719.
Morton JP, Karim SA, Graham K, Timpson P, Jamieson N, Athineos D et al. Dasatinib inhibits the development of metastases in a mouse model of pancreatic ductal adenocarcinoma. Gastroenterology 2010; 139: 292–303.
Erami Z, Herrmann D, Warren SC, Nobis M, McGhee EJ, Lucas MC et al. Intravital FRAP imaging using an E-cadherin-GFP mouse reveals disease-and drug-dependent dynamic regulation of cell-cell junctions in live tissue. Cell Rep 2016; 14: 152–167.
Antoni D, Burckel H, Josset E, Noel G . Three-dimensional cell culture: a breakthrough in vivo. Int J Mol Sci 2015; 16: 5517–5527.
Tian X, Liu Z, Niu B, Zhang J, Tan TK, Lee SR et al. E-cadherin/β-catenin complex and the epithelial barrier. J Biomed Biotechnol 2011; Article ID 567305.
Kalluri R, Weinberg RA . The basics of epithelial-mesenchymal transition. J Clin Invest 2009; 119: 1420–1428.
Kim K, Lu Z, Hay ED . Direct evidence for a role of beta-catenin/LEF-1 signaling pathway in induction of EMT. Cell Biol Int 2002; 26: 463–476.
Tsai JH, Yang J . Epithelial–mesenchymal plasticity in carcinoma metastasis. Genes & development 2013; 27: 2192–2206.
Mizuno S, Nakamura T . HGF–Met cascade, a key target for inhibiting cancer metastasis: The impact of NK4 discovery on cancer biology and therapeutics. Int J Mol Sci 2013; 14: 888–919.
Shamir ER, Pappalardo E, Jorgens DM, Coutinho K, Tsai WT, Aziz K et al. Twist1-induced dissemination preserves epithelial identity and requires E-cadherin. J Cell Biol 2014; 204: 839–856.
David JM, Rajasekaran AK . Dishonorable discharge: the oncogenic roles of cleaved E-cadherin fragments. Cancer Res 2012; 72: 2917–2923.
Nobis M, McGhee EJ, Morton JP, Schwarz JP, Karim SA, Quinn J et al. Intravital FLIM-FRET imaging reveals dasatinib-induced spatial control of src in pancreatic cancer. Cancer Res 2013; 73: 4674–4686.
Gargalionis AN, Karamouzis MV, Papavassiliou AG . The molecular rationale of Src inhibition in colorectal carcinomas. Int J Cancer 2014; 134: 2019–2029.
Al-Sohaily S, Henderson C, Selinger C, Pangon L, Segelov E, Kohonen-Corish MR et al. Loss of special AT-rich sequence-binding protein 1 (SATB1) predicts poor survival in patients with colorectal cancer. Histopathology 2014; 65: 155–163.
Kohonen-Corish MR, Tseung J, Chan C, Currey N, Dent OF, Clarke S et al. KRAS mutations and CDKN2A promoter methylation show an interactive adverse effect on survival and predict recurrence of rectal cancer. Int J Cancer 2014; 134: 2820–2828.
Pangon L, Ng I, Giry-Laterriere M, Currey N, Morgan A, Benthani F et al. JRK is a positive regulator of beta-catenin transcriptional activity commonly overexpressed in colon, breast and ovarian cancer. Oncogene 2016; 35: 2834–2841.
Giry-Laterriere M, Cherpin O, Kim YS, Jensen J, Salmon P . Polyswitch lentivectors: "all-in-one" lentiviral vectors for drug-inducible gene expression, live selection, and recombination cloning. Hum Gene Ther 2011; 22: 1255–1267.
Canel M, Serrels A, Miller D, Timpson P, Serrels B, Frame MC et al. Quantitative in vivo imaging of the effects of inhibiting integrin signalling via Src and FAK on cancer cell movement: effects on E-cadherin dynamics. Cancer Res 2010; 70: 9413–9422.
Howard S, Deroo T, Fujita Y, Itasaki N . A positive role of cadherin in wnt/β-catenin signalling during epithelial-mesenchymal transition. PLoS ONE 2011; 6: e23899.
Chen CL, Chen HC . Functional suppression of E-cadherin by protein kinase Cdelta. J Cell Sci 2009; 122: 513–523.
Acknowledgements
We thank Irvin Ng, Melissa Abas, Sabine Meessen, Monica Killen and Sean Warren for advice and technical assistance. The work was supported by Cancer Council NSW (RG 14-08, RG17-05), NHMRC (1020406, 1043501, 1089497, 1105640, 1129401), Cancer Institute NSW (10CDF232, 12CDF234), the Australian Research Council Future Fellowship (FT120100880), the Len Ainsworth Pancreatic Cancer Fellowship, Cancer Australia and Cure Cancer Australia (1049846), Gastroenterological Society of Australia, Sydney Catalyst and Tour de Cure. The contents of the published material are solely the responsibility of the administering institution, a participating institution or individual authors and do not reflect the views of the NHMRC. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Benthani, F., Herrmann, D., Tran, P. et al. ‘MCC’ protein interacts with E-cadherin and β-catenin strengthening cell–cell adhesion of HCT116 colon cancer cells. Oncogene 37, 663–672 (2018). https://doi.org/10.1038/onc.2017.362
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2017.362