Abstract
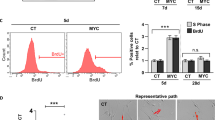
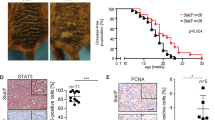
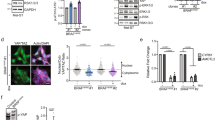
The MAPK pathway is activated in the majority of melanomas and is the target of therapeutic approaches. Under normal conditions, it initiates the so-called immediate early response, which encompasses the transient transcription of several genes belonging to the AP-1 transcription factor family. Under pathological conditions, such as continuous MAPK pathway overactivation due to oncogenic alterations occurring in melanoma, these genes are constitutively expressed. The consequences of a permanent expression of these genes are largely unknown. Here, we show that FOSL1 is the main immediate early AP-1 member induced by melanoma oncogenes. We first examined its role in established melanoma cells. We found that FOSL1 is involved in melanoma cell migration as well as cell proliferation and anoikis-independent growth, which is mediated by the gene product of its target gene HMGA1, encoding a multipotent chromatin modifier. As FOSL1 expression is increased in patient melanoma samples compared to nevi, we investigated the effect of enhanced FOSL1 expression on melanocytes. Intriguingly, we found that FOSL1 acts oncogenic and transforms melanocytes, enabling subcutaneous tumor growth in vivo. During the process of transformation, FOSL1 reprogrammed the melanocytes and downregulated MITF in a HMGA1-dependent manner. At the same time, AXL was upregulated, leading to a shift in the MITF/AXL balance. Furthermore, FOSL1 re-enforced pro-tumorigenic transcription factors MYC, E2F3 and AP-1. Together, this led to the enhancement of several growth-promoting processes, such as ribosome biogenesis, cellular detachment and pyrimidine metabolism. Overall, we demonstrate that FOSL1 is a novel reprogramming factor for melanocytes with potent tumor transformation potential.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Mandal R, Becker S, Strebhardt K . Stamping out RAF and MEK1/2 to inhibit the ERK1/2 pathway: an emerging threat to anticancer therapy. Oncogene 2016; 35: 2547–2561.
Vogt PK . Fortuitous convergences: the beginnings of JUN. Nat Rev Cancer 2002; 2: 465–469.
Lopez-Bergami P, Kim H, Dewing A, Goydos J, Aaronson S, Ronai Z . c-Jun regulates phosphoinositide-dependent kinase 1 transcription: implication for Akt and protein kinase C activities and melanoma tumorigenesis. J Biol Chem 2010; 285: 903–913.
Riesenberg S, Groetchen A, Siddaway R, Bald T, Reinhardt J, Smorra D et al. MITF and c-Jun antagonism interconnects melanoma dedifferentiation with pro-inflammatory cytokine responsiveness and myeloid cell recruitment. Nat Commun 2015; 6: 8755.
Kappelmann M, Kuphal S, Meister G, Vardimon L, Bosserhoff AK . MicroRNA miR-125b controls melanoma progression by direct regulation of c-Jun protein expression. Oncogene 2013; 32: 2984–2991.
Spangler B, Vardimon L, Bosserhoff AK, Kuphal S . Post-transcriptional regulation controlled by E-cadherin is important for c-Jun activity in melanoma. Pigment Cell Melanoma Res 2011; 24: 148–164.
Spangler B, Kappelmann M, Schittek B, Meierjohann S, Vardimon L, Bosserhoff AK et al. ETS-1/RhoC signaling regulates the transcription factor c-Jun in melanoma. Int J Cancer 2012; 130: 2801–2811.
Cohen DR, Curran T . fra-1: a serum-inducible, cellular immediate-early gene that encodes a fos-related antigen. Mol Cell Biol 1988; 8: 2063–2069.
Nishina H, Sato H, Suzuki T, Sato M, Iba H . Isolation and characterization of fra-2, an additional member of the fos gene family. Proc Natl Acad Sci USA 1990; 87: 3619–3623.
Tice DA, Soloviev I, Polakis P . Activation of the Wnt pathway interferes with serum response element-driven transcription of immediate early genes. J Biol Chem 2002; 277: 6118–6123.
Chinenov Y, Kerppola TK . Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene 2001; 20: 2438–2452.
Shaulian E, Schreiber M, Piu F, Beeche M, Wagner EF, Karin M . The mammalian UV response: c-Jun induction is required for exit from p53-imposed growth arrest. Cell 2000; 103: 897–907.
Ting CH, Chen YC, Wu CJ, Chen JY . Targeting FOSB with a cationic antimicrobial peptide, TP4, for treatment of triple-negative breast cancer. Oncotarget 2016; 7: 40329–40347.
Nakabeppu Y, Oda S, Sekiguchi M . Proliferative activation of quiescent Rat-1A cells by delta FosB. Mol Cell Biol 1993; 13: 4157–4166.
Saez E, Rutberg SE, Mueller E, Oppenheim H, Smoluk J, Yuspa SH et al. c-fos is required for malignant progression of skin tumors. Cell 1995; 82: 721–732.
Desmet CJ, Gallenne T, Prieur A, Reyal F, Visser NL, Wittner BS et al. Identification of a pharmacologically tractable Fra-1/ADORA2B axis promoting breast cancer metastasis. Proc Natl Acad Sci USA 2013; 110: 5139–5144.
Iskit S, Schlicker A, Wessels L . Peeper DS. Fra-1 is a key driver of colon cancer metastasis and a Fra-1 classifier predicts disease-free survival. Oncotarget 2015; 6: 43146–43161.
Yang S, Li Y, Gao J, Zhang T, Li S, Luo A et al. MicroRNA-34 suppresses breast cancer invasion and metastasis by directly targeting Fra-1. Oncogene 2013; 32: 4294–4303.
Renaud SJ, Kubota K, Rumi MA, Soares MJ . The FOS transcription factor family differentially controls trophoblast migration and invasion. J Biol Chem 2014; 289: 5025–5039.
Basbous J, Chalbos D, Hipskind R, Jariel-Encontre I, Piechaczyk M . Ubiquitin-independent proteasomal degradation of Fra-1 is antagonized by Erk1/2 pathway-mediated phosphorylation of a unique C-terminal destabilizer. Mol Cell Biol 2007; 27: 3936–3950.
Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE Jr et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet 2009; 41: 544–552.
Shain AH, Yeh I, Kovalyshyn I, Sriharan A, Talevich E, Gagnon A et al. The Genetic Evolution of Melanoma from Precursor Lesions. N Engl J Med 2015; 373: 1926–1936.
Vredeveld LC, Possik PA, Smit MA, Meissl K, Michaloglou C, Horlings HM et al. Abrogation of BRAFV600E-induced senescence by PI3K pathway activation contributes to melanomagenesis. Genes Dev 2012; 26: 1055–1069.
Widmer DS, Cheng PF, Eichhoff OM, Belloni BC, Zipser MC, Schlegel NC et al. Systematic classification of melanoma cells by phenotype-specific gene expression mapping. Pigment Cell Melanoma Res 2012; 25: 343–353.
Vincek V, Xu S, Fan YS . Comparative genome hybridization analysis of laser-capture microdissected in situ melanoma. J Cutan Pathol 2010; 37: 3–7.
Landsberg J, Kohlmeyer J, Renn M, Bald T, Rogava M, Cron M et al. Melanomas resist T-cell therapy through inflammation-induced reversible dedifferentiation. Nature 2012; 490: 412–416.
Leikam C, Hufnagel AL, Otto C, Murphy DJ, Muhling B, Kneitz S et al. In vitro evidence for senescent multinucleated melanocytes as a source for tumor-initiating cells. Cell Death Dis 2015; 6: e1711.
Thomas S, Thomas M, Wincker P, Babarit C, Xu P, Speer MC et al. Human neural crest cells display molecular and phenotypic hallmarks of stem cells. Hum Mol Genet 2008; 17: 3411–3425.
Belton A, Gabrovsky A, Bae YK, Reeves R, Iacobuzio-Donahue C, Huso DL et al. HMGA1 induces intestinal polyposis in transgenic mice and drives tumor progression and stem cell properties in colon cancer cells. PLoS One 2012; 7: e30034.
Shah SN, Kerr C, Cope L, Zambidis E, Liu C, Hillion J et al. HMGA1 reprograms somatic cells into pluripotent stem cells by inducing stem cell transcriptional networks. PLoS One 2012; 7: e48533.
Muller J, Krijgsman O, Tsoi J, Robert L, Hugo W, Song C et al. Low MITF/AXL ratio predicts early resistance to multiple targeted drugs in melanoma. Nat Commun 2014; 5: 5712.
Martin RM, Tunnemann G, Leonhardt H, Cardoso MC . Nucleolar marker for living cells. Histochem Cell Biol 2007; 127: 243–251.
Gentilella A, Kozma SC, Thomas G . A liaison between mTOR signaling, ribosome biogenesis and cancer. Biochim Biophys Acta 2015; 1849: 812–820.
Elkon R, Loayza-Puch F, Korkmaz G, Lopes R, van Breugel PC, Bleijerveld OB et al. Myc coordinates transcription and translation to enhance transformation and suppress invasiveness. EMBO Rep 2015; 16: 1723–1736.
Das KC, Muniyappa H . c-Jun-NH2 terminal kinase (JNK)-mediates AP-1 activation by thioredoxin: phosphorylation of cJun, JunB, and Fra-1. Mol Cell Biochem 2010; 337: 53–63.
Wellbrock C, Arozarena I . Microphthalmia-associated transcription factor in melanoma development and MAP-kinase pathway targeted therapy. Pigment Cell Melanoma Res 2015; 28: 390–406.
Bianchi-Smiraglia A, Bagati A, Fink EE, Moparthy S, Wawrzyniak JA, Marvin EK et al. Microphthalmia-associated transcription factor suppresses invasion by reducing intracellular GTP pools. Oncogene 2016; 36: 84–96.
Lister JA, Capper A, Zeng Z, Mathers ME, Richardson J, Paranthaman K et al. A conditional zebrafish MITF mutation reveals MITF levels are critical for melanoma promotion vs. regression in vivo. J Invest Dermatol 2014; 134: 133–140.
Ploper D, Taelman VF, Robert L, Perez BS, Titz B, Chen HW et al. MITF drives endolysosomal biogenesis and potentiates Wnt signaling in melanoma cells. Proc Natl Acad Sci USA 2015; 112: E420–E429.
Sensi M, Catani M, Castellano G, Nicolini G, Alciato F, Tragni G et al. Human cutaneous melanomas lacking MITF and melanocyte differentiation antigens express a functional Axl receptor kinase. J Invest Dermatol 2011; 131: 2448–2457.
Tirosh I, Izar B, Prakadan SM, Wadsworth MH 2nd, Treacy D, Trombetta JJ et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016; 352: 189–196.
Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA et al. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev 2002; 16: 245–256.
Kapoor A, Yao W, Ying H, Hua S, Liewen A, Wang Q et al. Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell 2014; 158: 185–197.
Zanconato F, Forcato M, Battilana G, Azzolin L, Quaranta E, Bodega B et al. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat Cell Biol 2015; 17: 1218–1227.
Huang, da W, Sherman BT, Lempicki RA . Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4: 44–57.
Acknowledgements
This work was supported by the Melanoma Research Network of the Deutsche Krebshilfe eV (German Cancer Aid) and by the research unit FOR2314, project 5 (German Research Foundation). We are furthermore grateful to Manfred Gessler (Dept of Developmental Biochemistry, University of Würzburg) for providing us with the pSB-ET-iE vector and to Stefan Gaubatz (Dept. of Physiological Chemistry, University of Würzburg) for the E2F3 antibody. We would also like to thank Marie-Christine Dabauvalle (Dept of Zoology and Developmental Biology, University of Würzburg) for sharing the fibrillarin antibody with us.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Rights and permissions
About this article
Cite this article
Maurus, K., Hufnagel, A., Geiger, F. et al. The AP-1 transcription factor FOSL1 causes melanocyte reprogramming and transformation. Oncogene 36, 5110–5121 (2017). https://doi.org/10.1038/onc.2017.135
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2017.135
This article is cited by
-
Multiple Fra-1-bound enhancers showing different molecular and functional features can cooperate to repress gene transcription
Cell & Bioscience (2023)
-
Illuminating phenotypic drug responses of sarcoma cells to kinase inhibitors by phosphoproteomics
Molecular Systems Biology (2023)
-
Age-related self-DNA accumulation may accelerate arthritis in rats and in human rheumatoid arthritis
Nature Communications (2023)
-
The nuclear oncoprotein Fra-1: a transcription factor knocking on therapeutic applications’ door
Oncogene (2020)
-
FOSL1 is a novel mediator of endotoxin/lipopolysaccharide-induced pulmonary angiogenic signaling
Scientific Reports (2020)