Abstract
The expression of immediate early response 3 (IER3), a protein with a short half-life, is rapidly induced by various cellular stimuli. We recently reported that IER3 induces the apoptosis of cervical cancer cells and that its expression is downregulated in patients with cervical cancer. However, the molecular mechanism involved in the rapid degradation of IER3 remains unknown. Here, we demonstrate that MDM2 is an E3 ligase that interacts with IER3 and promotes its ubiquitination, followed by proteasomal degradation. Polyubiquitination of the conserved lysine 60 of IER3 is essential for its degradation. In addition, four and a half LIM domains protein 2 (FHL2) binds to both IER3 and MDM2, allowing for efficient MDM2-mediated IER3 degradation by facilitating an association between MDM2 and IER3. Moreover, IER3 induces cell cycle arrest in cervical cancer cells and its activity is further enhanced in cells in which FHL2 or MDM2 was silenced, thereby preventing IER3 degradation. The E6 and E7 oncoproteins of human papilloma virus 18 regulated IER3 expression. FHL2 expression was significantly higher in the squamous epithelium of cervical carcinoma tissues than in non-cancerous cervical tissues, whereas cervical carcinoma expression of IER3 was downregulated in this region. Thus, we determined the molecular mechanism responsible for IER3 degradation, involving a ternary complex of IER3, MDM2 and FHL2, which may contribute to cervical tumor growth. Furthermore, we demonstrated that FHL2 serves as a scaffold for E3 ligase and its substrate during the ubiquitination reaction, a function that has not been previously reported for this protein.
Similar content being viewed by others
Introduction
Regulation of protein degradation is a fundamental homeostasis mechanism that controls protein levels in cells.1 In eukaryotes, intracellular protein degradation is achieved by ubiquitin-mediated proteasomal destruction and lysosome-mediated proteolysis.2 The ubiquitin-proteasomal pathway constitutes a selective process in which the concerted actions of an ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2) and ubiquitin-protein ligase (E3) lead to the attachment of ubiquitin to the lysine residues of substrate proteins, and the product is then recognized by the 26S proteasome.3, 4 During this process, E3 ligases play a central role as they recognize specific protein substrates and catalyze ubiquitin transfer.5 MDM2 is a RING finger family E3 ubiquitin ligase proto-oncogene known for mediating the degradation of the tumor-suppressor p53.6, 7
The expression of immediate early response gene 3 (IER3) is rapidly induced by various cellular stimuli such as DNA damage, growth factors, cytokines and viral infection.8, 9 IER3 regulates cellular apoptosis, proliferation, DNA repair, differentiation and inflammation.10, 11, 12, 13, 14 We recently reported that IER3 induces apoptosis in cervical cancer cells and its expression is downregulated in tissues from patients with cervical carcinoma.15 IER3 is known to have a short half-life and its degradation can be prevented by a proteasomal inhibitor.16 However, the molecular mechanism involved in the regulation of IER3 degradation remains unknown.
Four and a half LIM domains protein 2 (FHL2), also known as DRAL, belongs to the four and a half LIM domains protein family.17, 18 FHL2 is a multifunctional protein involved in a wide range of cellular processes, including the regulation of gene expression, cell survival, proliferation, differentiation, adhesion and motility.19, 20, 21 Because of its structural nature, FHL2, consisting of multiple LIM motifs that serve as protein-binding sites, interacts with diverse types of proteins and assembles multi-protein complexes.20, 22
Cervical cancer is the second leading cause of cancer-related death in women worldwide.23 Infection with human papilloma viruses (HPVs), including HPV 16 and 18, is the major etiology for cervical malignancy.24, 25 Viral E6 and E7 from high-risk HPVs are two critical oncoproteins for cervical cancer development by deregulation of cell proliferation, apoptosis and genome instability.26 Although little is known regarding the role of FHL2 in cervical cancer, its interaction with E6 and E7 oncoproteins has been reported.27, 28
In this study, we revealed the signaling mechanism involved in IER3 degradation. We demonstrated that IER3 undergoes MDM2-mediated polyubiquitination followed by proteasomal degradation, in which FHL2 acts as a scaffold that facilitates the association of MDM2 to IER3. In addition, we found that IER3 induces cell cycle arrest in cervical carcinoma cells, which is further increased in either FHL2- or MDM2-depleted cells. Furthermore, a significant upregulation of FHL2 and downregulation of IER3 in the squamous epithelium of cervical carcinoma tissues suggest that the FHL2-mediated IER3 degradation could be implicated in the development of cervical carcinoma.
Results
FHL2 interacts with IER3
To understand the intracellular molecular mechanism involved in the regulation of IER3, yeast two-hybrid screening of a human ovarian cDNA library was performed using the full-length human IER3 as the bait. Among isolated interacting clones, three were identified as FHL2 and the specific interaction between IER3 and FHL2 was confirmed in the yeast two-hybrid system (Figure 1a). The in vivo interaction of these two proteins was verified by immunoprecipitation followed by western blot analysis after overexpression in 293T cells (Figures 1b and c). In addition, the association of endogenous FHL2 and IER3 proteins was observed in HeLa cervical carcinoma cells (Figure 1d). Immunofluorescence confocal microscopic analysis showed that endogenous FHL2 and IER3 were co-localized in the cytoplasm of HeLa cells (Figure 1e).
Identification of FHL2 as a novel interacting protein of IER3. (a) Yeast growth was demonstrated in colonies expressing both FHL2 and IER3 fused to the GAL4 DNA activation and binding domains, respectively. (b, c) 293T cells were co-transfected with Myc-FHL2 and FLAG-IER3 (b) or HA-FHL2 and FLAG-IER3 (c). After 24 h of incubation, cell lysates were prepared and immunoprecipitated with anti-FLAG (b) or anti-HA (c) antibodies. Immunoblot analyses were performed using the indicated antibodies. (d) The interaction between endogenous FHL2 and IER3 proteins was determined in HeLa cells after immunoprecipitation with control IgG or an anti-IER3 antibody. For all immunoblot images presented throughout this manuscript, the membrane was cut into pieces according to the estimated molecular weight of the proteins of interest and probed with the indicated antibodies. All cropped blots were run under the same experimental conditions. (e) Intracellular co-localization of endogenous FHL2 and IER3 proteins in HeLa cells was assessed by fluorescence confocal microscopy.
The association of FHL2 and IER3 is mediated by LIM3 and 4 domains and the PEST (proline, glutamic acid, serine, threonine)-rich region
To define the binding domain that mediates the interaction of FHL2 and IER3, we generated plasmids encoding HA-tagged full-length and deleted mutant forms of FHL2 (Figure 2a). These constructs were co-transfected with glutathione sepharose (GST)-tagged IER3 into HeLa cells and then immunoprecipitated. As shown in Figure 2b, FHL2 mutants lacking the LIM4 and/or LIM3 domains at the C-terminal end (ΔC1 and ΔC2) failed to bind to IER3, indicating that the these LIM domains are involved for their association. We also produced plasmids encoding GST-tagged full-length and serially deleted mutants of IER3 (Figure 2c). The IER3 mutant with deleted sequences from amino acid 26 to 50 (Δ2) did not interact with FHL2, suggesting that this PEST-rich region mediates the binding to FHL2 (Figure 2d). Furthermore, we generated additional mutants of FHL2 (ΔN3 and ΔN4) and IER3 (PEST) to determine whether the minimal binding motives mapped from Figures 2b and d are sufficient for their interaction. As shown in Figure 2e, the LIM 3 and 4 domains of FHL2 and the PEST-rich region (amino acids 26–55) of IER3 were sufficient for their association.
Mapping of the binding regions for the interaction between FHL2 and IER3. (a) Structures of the plasmids encoding HA-tagged full-length and truncated mutants of FHL2 are illustrated. (b) HeLa cells were co-transfected with each HA-tagged FHL2 construct and GST-IER3. Twenty-four hours after transfection, cell lysates were prepared and immunoprecipitated with GST beads. Arrows indicate expected positions of FHL2 proteins. GAPDH was included as a loading control. (c) Structures of GST-tagged full-length and deletion mutants of IER3 are illustrated. (d) Immunoprecipitation was performed as described in b following transfection with GST-IER3 constructs and Myc-FHL2 into HeLa cells. Arrows indicate IER3 proteins. (e) Transfection and immunoprecipitation were performed as described in b with indicated DNA constructs. Arrows indicate expected sizes of FHL2 mutants.
FHL2 stimulates ubiquitination-mediated proteasomal degradation of IER3
To investigate the functional role of the association between FHL2 and IER3, these two proteins were overexpressed in HeLa cells. The IER3 protein level was decreased by FHL2 overexpression (Figure 3a). FHL2 knockdown using specific small interfering RNAs (siRNAs) increased the levels of endogenous IER3 (Figure 3b and Supplementary Figure 1a) as well as its stability (Figure 3c). In contrast, the IER3 mRNA level was not affected by the modulation of FHL2 expression, as determined by quantitative real-time PCR analysis (Supplementary Figure 2). Cellular proteins are mainly degraded via the ubiquitin-mediated proteasomal and lysosomal pathways.2 Thus, we determined the degradation pathway involved in FHL2-induced downregulation of IER3 expression using inhibitors of the two degradation pathways. FHL2-induced downregulation of IER3 was completely blocked by the proteasomal inhibitor, MG132, but not by the lysosomal inhibitor, chloroquine (Figure 3d). In addition, IER3 underwent ubiquitination, which was augmented by FHL2 overexpression, whereas the FHL2 mutant lacking IER3-binding capacity (ΔC2) failed to increase IER3 ubiquitination (Figure 3e). The attenuation of IER3 ubiquitination in FHL2-depleted cells further supports that FHL2 stimulates IER3 polyubiquitination in cells (Figure 3f). A decrease in endogenous IER3 ubiquitination was also confirmed in FHL2 knockdown HeLa cells (Figure 3g).
Regulation of IER3 stability by FHL2 via the promotion of ubiquitination-mediated proteasomal degradation. (a) HeLa cells were transfected with FLAG-IER3 in the presence or absence of HA-FHL2. The pEGFP plasmid (300 ng) was co-transfected as an internal control of transfection efficiency. Twenty-four hours after transfection, cell lysates were prepared and subjected to SDS–polyacrylamide gel electrophoresis. The IER3 level was determined by western blot analysis. (b) Immunoblot (IB) results from HeLa cells after knockdown of FHL2 by FHL2 siRNA (100 and 200 nM) transfection are shown. (c) Twenty-one hours after transfection of HeLa cells with FHL2 siRNA, the cells were incubated with cycloheximide (CHX; 100 μg/ml), harvested at the indicated time points and analyzed by IB. Quantitative analysis of IER3 levels from three independent experiments is presented (lower panel). Asterisks indicate statistically significant differences (P<0.05). (d) HeLa cells were co-transfected with FLAG-IER3 and HA-FHL2 or the pCMV-HA empty vector. Twelve hours after transfection, the cells were treated with dimethylsulphoxide (0.1%), MG132 (50 μM) or chloroquine (50 μM) for 12 h. The cell lysates were subjected to IB analysis. (e, f) HeLa cells were transfected with FLAG-IER3 and HA-ubiquitin together with HA-FHL2 or HA-FHL2ΔC2 (e), or FHL2 siRNA (f). Twelve hours after transfection, the cells were incubated with MG132 (50 μM) for 12 h followed by immunoprecipitation (IP) and IB analysis. Arrows indicate the expected position of mono-, di- and tri-ubiquitinated IER3. (g) HeLa cells were transfected with siRNA for FHL2. Twelve hours after transfection, the cells were incubated with MG132 (50 μM) for 12 h followed by IP and IB analysis.
Lysine (K) 60 residue is the primary ubiquitination site on IER3
Human IER3 possesses only two lysine residues. Comparative sequence analysis of IER3 orthologs in mammalian species revealed two conserved lysine residues at the amino-acid positions 60 and 84 (Figure 4a). We generated two IER3 mutants in which the K residues were replaced with arginine (R) residues, K60R and K84R, and compared their protein stabilities. Although the levels of both wild-type (WT)-IER3 and the K84R mutant increased in the presence of MG132, the K60R mutant protein was present at a higher basal level, which was not affected by the proteasome inhibitor (Figure 4b). Accordingly, the levels of WT and K84R IER3 were clearly increased by FHL2 knockdown, whereas the level of the K60R mutant did not change (Figure 4c). Moreover, FHL2 overexpression failed to decrease the level of K60R IER3 (Figure 4d). Consistently, K60R IER3 was resistant to ubiquitination, even after forced expression of FHL2 (Figure 4e). This lack of ubiquitination was not a consequence of the inability of IER3 to interact with FHL2, as these proteins showed efficient binding (Figure 4e).
Identification of IER3 lysine 60 as the ubiquitin acceptor site. (a) The alignment of amino-acid sequences of mammalian IER3 proteins encompassing two lysines, K60 and K84 (red), is shown. (b) The stability of the GST-tagged IER3 wild-type (WT) and substitution mutants was determined after transfection of HeLa cells with the respective plasmids. Twelve hours after transfection, the cells were incubated with MG132 (50 μM) for 12 h, and cell lysates were analyzed by western blot analysis. (c, d) Changes in the protein stability of IER3 WT and mutants in response to FHL2 knockdown (c) or overexpression (d) were assessed by western blot analysis. A green fluorescent protein (GFP)-expressing plasmid was included as an inner control of transfection efficiency. (e) An in vivo ubiquitination assay was performed after transfection of HeLa cells with the indicated plasmids followed by immunoprecipitation.
IER3 is a novel substrate of MDM2 E3 ligase
To identify the specific E3 ubiquitin ligase targeting IER3, the interaction of E3 ligases with IER3 was examined by immunoprecipitation. We found that IER3 associated with MDM2, but not with other E3 ligases such as SKP2 and β-TrCP (Figure 5a). Immunoblot analysis of HeLa cell lysates also showed that MDM2 overexpression prominently decreased the IER3 level (Figure 5a). A direct interaction between IER3 and MDM2 was also demonstrated by immunoprecipitation using purified recombinant proteins (Figure 5b and Supplementary Figure 3a). To determine the binding region, truncated MDM2 mutants were generated (Figure 5c). Immunoprecipitation results shows that the ring domain of MDM2 mediated its interaction with IER3, as the ΔC1 and ΔC2 mutants failed to associate with IER3 (Figure 5d).
Identification of MDM2 as the E3 ubiquitin ligase for IER3. (a) The interaction of IER3 with different E3 ubiquitin ligases was determined by immunoprecipitation (IP) in HeLa cells. The arrows indicate the expected positions of the proteins. (b) In vitro, a direct interaction between IER3 and MDM2 was determined by IP using recombinant GST-IER3 (1 μg) and MDM2 (0.5 μg) proteins. (c) Structures of GST-tagged full-length and truncated mutants of MDM2 are illustrated. (d) HeLa cells were co-transfected with each GST-tagged MDM2 construct and FLAG-IER3. Twenty-four hours after transfection, cell lysates were prepared and IP with GST beads. The arrows indicate the expected positions of MDM2 proteins. (e) MDM2 was knocked down in HeLa cells using MDM2 siRNA (100 and 200 nM) and the cell lysates were analyzed by immunoblotting (IB). (f) HeLa cells were transfected with the indicated plasmids and changes in the stability of IER3 proteins induced by MDM2 were determined by IB analysis. Green fluorescent protein (GFP)-expressing plasmid was included as an internal control of transfection efficiency. (g) Degrees of IER3 ubiquitination (Ub) were determined by IP after HeLa cells were transfected with GST-IER3 and HA-ubiquitin together with the MDM2 plasmid. Twelve hours after transfection, the cells were incubated with MG132 (50 μM) for 12 h followed by IP and IB analysis. (h) MDM2 was knocked down using a siRNA against MDM2 in HeLa cells. Twelve hours after transfection, the cells were incubated with MG132 (50 μM) for 12 h followed by IP and IB analysis. (i) In vitro Ub of IER3 by MDM2 was assessed by the incubation of recombinant IER3 protein (1 μg) in Ub buffer containing MDM2 protein (0.3 μg). The western blot results are shown.
MDM2 knockdown increased the level of endogenous IER3 without affecting the level of FHL2 (Figure 5e and Supplementary Figure 1b). This upregulation of IER3 upon the knockdown of either FHL2 or MDM2 was also observed in different cervical carcinoma cells, SiHa cells (Supplementary Figure 4). MDM2 overexpression did not decrease the level of the ubiquitination-resistant K60R mutant (Figure 5f). Enhanced production of polyubiquitinated IER3 following MDM2 overexpression (Figure 5g) and attenuated ubiquitination of endogenous IER3 in MDM2-silenced HeLa cells (Figure 5h) were confirmed. Moreover, an in vitro ubiquitination assay using recombinant IER3 and MDM2 proteins also demonstrated the induction of IER3 ubiquitination by MDM2 (Figure 5i).
FHL2 stimulates MDM2-mediated ubiquitination of IER3 by forming a ternary complex
Next, we investigated the inter-regulatory mechanism that promotes IER3 degradation by FHL2 and MDM2. FHL2-mediated downregulation of IER3 was prevented by silencing of MDM2 (Figure 6a), indicating that the stimulating effect of FHL2 on IER3 proteasomal degradation involves the action of MDM2. Similarly, MDM2 failed to downregulate IER3 when FHL2 was silenced (Figure 6b), suggesting that FHL2 is necessary for MDM2 activity to degrade IER3. We found that FHL2 also directly interacts with MDM2 as determined by the in vitro pull-down assay using recombinant FHL2 and GST-MDM2 proteins (Figure 6c). Their interaction was mediated by the LIM2 domain of FHL2 (Figure 6d) and the region (amino acids 101–234) between the p53-binding motif and acidic domain of MDM2 (Figure 6e). This ternary complex formation by endogenous proteins was further confirmed in a two-step immunoprecipitation experiment (Supplementary Figure 5a).
Regulation of MDM2-mediated degradation of IER3 by FHL2. (a, b) Changes in IER3 levels were determined by western blot analysis in HeLa cells after transfection with the indicated plasmids and siRNAs (200 nM). (c) The direct interaction between FHL2 and MDM2 was assessed by an in vitro immunoprecipitation (IP) after incubation of recombinant FHL2 (1 μg) and GST-MDM2 (0.5 μg) proteins. (d) The HA-tagged WT and mutant FHL2 constructs shown in Figure 2a were co-transfected with MDM2 into HeLa cells and their binding capacities were assessed by IP. The arrows indicate the expected positions of FHL2 proteins. (e) The GST-tagged WT and mutant MDM2 constructs presented in Figure 5c were co-transfected with Myc-FHL2 into HeLa cells and their binding capacities were assessed by IP. The arrows indicate the expected positions of MDM2 proteins. (f, g) The effect of FHL2 expression on the association of IER3 and MDM2 was determined after transfection of HeLa cells with the indicated plasmids and siRNA followed by IP and western blot analysis. The relative interaction ratios are graphically presented from the results of three independent experiments (P<0.05). (h) The interaction between endogenous IER3 with endogenous FHL2 and MDM2 proteins were determined in HeLa cells after IP with a BS3-crosslinked anti-MDM2 antibody or control IgG. (i) Formation of a ternary complex in vitro was determined after incubation of 1 μg of recombinant proteins (FHL2, MDM2 and GST-IER3) followed by IP and western blot analysis. (j) A direct effect of FHL2 on the MDM2-mediated IER3 ubiquitination was assessed in an in vitro ubiquitination assay. Purified recombinant FHL2 (1 μg) and GST-IER3 (1 μg) proteins were incubated in Ub buffer containing MDM2 protein (0.3 μg). IB, immunoblot.
To address how FHL2 stimulates MDM2-induced IER3 degradation, the effect of FHL2 on the binding capacity of MDM2 to IER3 was determined by immunoprecipitation. FHL2 overexpression enhanced the interaction between MDM2 and IER3 by more than 2.5-fold, whereas overexpression of the FHL2 mutant lacking the ability to bind IER3 (ΔC2) did not affect the strength of the MDM2–IER3 interaction (Figure 6f). In addition, the interaction between MDM2 and IER3 was decreased in FHL2 knockdown HeLa cells (Figure 6g), indicating that FHL2 allows for the efficient binding of MDM2 to IER3. In addition, the formation of an endogenous ternary complex of FHL2-IER3-MDM2 in HeLa cells (Figure 6h and Supplementary Figure 5b) and a clearly increased in vitro association between MDM2 and IER3 in the presence of FHL2 recombinant protein (Figure 6i and Supplementary Figure 3b) were confirmed. Furthermore, the in vitro ubiquitination assay results confirmed that MDM2-mediated ubiquitination of IER3 was augmented by the addition of FHL2 protein (Figure 6j), and ubiquitinated IER3 was decreased by knockdown of either FHL2 or MDM2 (Supplementary Figure 6).
FHL2 and MDM2 regulate IER3-induced anti-proliferative activity
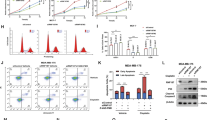
FHL2 overexpression promoted cell cycle progression to the S and G2/M phases, whereas the overexpression or knockdown of IER3 induced G0/G1 phase arrest or cell cycle progression, respectively (Figures 7a and b). In addition, the knockdown of either FHL2 or MDM2, which increases IER3 stability, decreased the cell population in the S phase. The IER3-induced inhibitory activity on the cell cycle progression to S phase was further increased in FHL2- or MDM2-silenced cells (Figure 7c). Simultaneously, consistent effects on cell proliferation were also observed by measuring 5′-bromo-2′-deoxyuridine-positive cells (Supplementary Figures 7a and b). Moreover, the regulatory roles of MDM2 and FHL2 on IER3 was further determined using mouse knockout cells of MDM2+/+/p53−/− and MDM2−/−/p53−/−. As shown in Figure 7d, FHL2 overexpression efficiently stimulated IER3 degradation in MDM2+/+/p53−/− cells but not in MDM2−/−/p53−/− cells. Accordingly, the anti-proliferative activity of IER3 was inhibited by FHL2 expression in MDM2-expressing cells but not in MDM2-null cells (Figure 7e).
Inhibition of IER3-induced cell cycle arrest by FHL2 and MDM2. Effects of FHL2 and IER3 on the cell cycle regulation were assessed after transfecting HeLa cells with IER3- or FHL2-encoding plasmids (a) and specific siRNA for IER3 (b). Twenty-four hours post transfection, the cells were harvested and analyzed by flow cytometry. Representative DNA content histograms are presented (left panel). Cell populations in the S phase were graphically presented as % of control. Asterisks indicate statistically significant differences from control values determined after transfection with an empty vector (P<0.05). (c) HeLa cells were transfected with the IER3 plasmid with or without siRNAs specific for MDM2 or FHL2 for 24 h and cell cycle phases were analyzed (top panel). Efficient silencing of FHL2 and MDM2 using specific siRNAs (200 nM) was confirmed by western blot analysis and is presented in the bottom panel. Different letters denote statistically significant differences (P<0.05). Data are presented as the mean±s.e.m. from three independent experiments performed in duplicate. (d, e) MDM2+/+/p53−/− and MDM2−/−/p53−/− MEF cells were transfected with FLAG-IER3 in the presence or absence of HA-FHL2. Twenty-four hours after transfection, immunoblotting (d) and 5′-bromo-2′-deoxyuridine cell proliferation assays (e) were performed. Different letters denote statistically significant differences (P<0.01). Data are presented as the mean±s.e.m. of three independent experiments performed in triplicate. (f) HeLa cells were transfected with siRNAs for HPV18 E6 or E7 (200 nM). Twenty-four hours after transfection, cell lysates were prepared and immunoblotted with the indicated antibodies. (g) HeLa cells were transfected with siRNAs for HPV18 E6 and/or MDM2 (200 nM) for 24 h. The cell lysates were analyzed by immunoblotting with the indicated antibodies.
Interestingly, MDM2 retained its ability to downregulate IER3 in p53-null cells (MDM2+/+/p53−/− vs MDM2−/−/p53−/−; Figure 7d). Accordingly, IER3 was still able to inhibit the proliferation of these p53-null cells (Figure 7e). This p53-independent regulation of IER3 was further confirmed in p53-knockdown HeLa cells (Supplementary Figure 7c), in which MDM2-mediated IER3 expression and the anti-proliferative function of IER3 was dependent on MDM2 but independent of p53.
As the E6 and E7 oncoproteins are critical in cervical cancer development, E6 and E7 were knocked down in HeLa cells, and the levels of IER3, FHL2 and MDM2 were assessed. As shown in Figures 7f and g, E6 knockdown resulted in the upregulation of MDM2 with concomitant downregulation of IER3 protein, whereas E7 knockdown induced the downregulation of IER3 without affecting the MDM2 level. The results of quantitative real-time PCR analysis suggest that E7 knockdown-induced IER3 downregulation occurs at the transcriptional level (Supplementary Figure 8).
The squamous epithelium of cervical carcinoma tissues exhibits inverse expression profiles for FHL2 and IER3
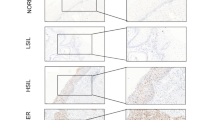
We further examined the expression profile of FHL2, IER3 and MDM2 in human cervical carcinoma tissues. For this purpose, a tissue microarray (TMA) containing cervical carcinoma and non-cancerous cervical tissues was constructed (Supplementary Table 1). Analysis of the FHL2 immunostaining of the TMA indicated a higher proportion of positive cells and stronger staining intensity in squamous epithelial cells of cervical carcinoma tissues relative to that observed in the non-cancerous cervical tissues (Figure 8a and Supplementary Figures 9a and d). The opposite expression pattern was observed when analyzing the IER3 immunostaining (Figure 8a and Supplementary Figures 9b and d). In contrast, no significant difference was observed in term of MDM2 expression between the two groups (Figure 8a and Supplementary Figures 9c and d). Representative images of IHC of each protein in higher magnification are presented in Supplementary Figure 9d. The receiver operating characteristic (ROC) curve analysis of FHL2 immunostaining showed perfect specificity (1.0) and sensitivity (1.0) to cervical carcinoma tissues in the TMA training set (Figure 8b and Supplementary Table 2). In addition, the ROC for IER3 immunostaining showed a good specificity (0.92) and sensitivity (0.86–0.78) to discriminate normal cervix and cervical carcinoma tissues, whereas MDM2 immunostaining exhibited no meaningful value (Figure 8b and Supplementary Table 2). ROC curve analysis of the proportion score (PS), intensity score (IS) and total scores of the TMA assay results, calculated as the sum of the PS (0–5) of positive cells and the IS (0–3 scales) of immunostaining, yielded area under the curves similar to those determined from the percentage of positive cells (Supplementary Table 2). Based on our findings, we proposed that the molecular regulation of IER3 degradation involves the interaction of FHL2 and MDM2 proteins, leading to the ubiquitination-mediated proteasomal degradation of IER3 (Figure 8c).
Upregulated FHL2 and downregulated IER3 in squamous epithelium of cervical cancer tissues. (a) Scatter histograms of the score determined by immunohistochemical staining of FHL2, IER3 and MDM2 are presented. Statistical differences in immunostaining analysis between cervical carcinoma (CA) and non-cancer (NC) cervical tissues in the TMA performed by two-tailed t-tests are shown. (b) ROC curve analysis of the proportion scores (PS), intensity scores (IS), and total scores (TSs) shows similar AUC (area under the curve) values. TSs were calculated as the sum of the PS in the range of 0–5 and the IS within the range of 0–3. (c) A proposed molecular mechanism involved in the regulation of IER3 degradation by FHL2 and MDM2 is presented.
Discussion
Considering that IER3 is an immediate early response protein having a short half-life, cells must possess a specific and rapid degradation machinery capable of such a tight control of IER3 expression. However, mechanisms underlying IER3 degradation remain unknown. Here, for the first time, we demonstrate that IER3 degradation is regulated by a multimeric protein complex that incorporates FHL2, MDM2 and IER3. The ubiquitin-mediated proteasome pathway mediates the targeted degradation of most short-lived proteins in eukaryotic cells by tagging proteins on lysine residues with ubiquitin.2 In this study, we identified MDM2 is the E3 ligase for IER3 and demonstrated that IER3 undergoes MDM2-mediated ubiquitination on the highly conserved K60 residue followed by proteasomal degradation.
The LIM domain consists of two zinc finger motifs, which mediate protein–protein interactions.22 FHL2 pleiotropic functions are likely attributable to the presence of multiple LIM domains. Here, we identified two novel FHL2-interacting proteins, IER3 and MDM2, which form a ternary complex. Of particular interest, FHL2 was necessary for the interaction of MDM2 ligase with IER3. Thus, we defined a unique and critical role of FHL2 in the regulation of IER3 protein degradation in which FHL2 is likely facilitating the efficient ubiquitination of IER3 through positioning IER3 and MDM2 in close proximity by binding to both proteins. These observations indicate that FHL2 functions as a scaffold or an adaptor protein required to convey the effective signal transduction involved in IER3 degradation.
The transforming activity of HPV infection is largely mediated by the HPV-derived E6 and E7 oncoproteins that dysregulate crucial signaling pathways.24, 29 The E6 protein binds to E6-associated protein and degrades p53, a well-known tumor suppressor, leading to increased cell cycle progression and inhibited apoptosis followed by the accumulation of DNA-damaged cells.29, 30 The E7 protein inactivates the tumor-suppressor family of retinoblastoma proteins, inducing uncontrolled activation of E2F transcriptional factor, consequently stimulating cell cycle progression.31, 32, 33 In the present study, we found that both E6 and E7 regulate IER3 expression in HPV-infected cells. IER3 downregulation induced by E6 depletion is likely the outcome of robust expression of MDM2, which facilitates IER3 degradation, whereas its downregulation by E7 depletion is likely associated with the transcriptional regulation of IER3 mRNA. Although additional studies exploring the precise mechanism are required, this finding further supports the tumor-suppressive role of IER3 in HPV-mediated carcinogenesis.
We previously reported that the p53 family member TAp73β is a specific transcriptional activator of IER3 in cervical cancer cells and defective expression of TAp73β in cervical carcinoma contributes to the downregulation of IER3 transcription.15 The current findings showing the FHL2-mediated enhanced IER3 protein degradation in cervical cancer cells and the inverse expression profiles of FHL2 and IER3 in cervical cancer tissues indicate the presence of an IER3 downregulation machinery at its protein level as well. Taken together, deficient expression of IER3 in cervical carcinoma and its critical role as an effective inducer of both apoptotic cell death15 and cell cycle arrest in cervical carcinoma cells (Figure 7) imply that IER3 is a crucial molecule, which possibly functions as a tumor suppressor during cervical cancer development. Therefore, based on these observations, we postulate that the inhibition of IER3 activity, resulting from IER3 downregulation at both the transcriptional and post-translational level, likely contributes to the growth of cervical carcinoma. In addition, our study demonstrates that IER3 expression is under a tight regulatory control to maintain cellular homeostasis and its perturbation may be linked to cervical tumor development and progression.
Cumulative evidence suggests that FHL2 function depends on the cell type and the interacting proteins.34 FHL2 expression profile also varies depending on the cancer type, it can be overexpressed or downregulated.34 In addition, FHL2 opposite activities, that is, the induction or inhibition of apoptosis and proliferation, have been reported in different cell types.35, 36, 37, 38, 39 Likewise, dual proapoptotic and antiapoptotic functions of IER3 that depend on the cellular context and contrasting IER3 expression profiles have been reported depending on the cancer type.40 Although the underlying cellular mechanism that accounts for these differences is still elusive, the interplay between FHL2 and IER3 may be implicated in these cell context-dependent contrasting outcomes.
Disturbed protein degradation is involved in multiple pathophysiological conditions, including cancer.41, 42 Proteasome inhibitors such as bortezomib have emerged as new chemotherapeutic agents.43, 44 Targeting and intervening with cell-type-specific molecules that regulate proteasomal degradation such as FHL2 would be an attractive and beneficial strategy to minimize side effects and, thus, the future application of FHL2 modulatory approaches is warranted. In addition, FHL2 immunostaining can serve as a good diagnostic tool for cervical cancer as it showed perfect specificity and sensitivity in the TMA (Figure 8b). In summary, we demonstrated a molecular network involved in the proteasome-mediated IER3 degradation, involving the ternary complex, FHL2–MDM2–IER3. This protein complex, where FHL2 has a key intermediary role, may be implicated in the development of cervical carcinoma.
Materials and methods
Mammalian cell culture and reagents
HeLa (American Type Culture Collection, Manassas, VA, USA), SiHa (Korean Cell Line Bank, Seoul, South Korea) and 293T (American Type Culture Collection) cells were cultured in Dulbecco's modified Eagle medium containing 10% fetal bovine serum and 1% penicillin–streptomycin. Cells were grown in an incubator at 37 °C with 5% CO2. Reagents used for cell culture were purchased from Caisson (Caisson, North Logan, UT, USA). MDM2+/+/p53−/− and MDM2−/−/p53−/− MEF cells were generous gifts from Dr Guillermina Lozano (University of Texas MD Anderson Cancer Center). The proteasome inhibitor MG132 was purchased from BioVison (Mountain View, CA, USA). Bis(sulfosuccinimidyl)suberate (BS3), a crosslinker, was purchased from Thermo Scientific (Rockford, IL, USA). Other agents, including chloroquine, cycloheximide and thrombin were purchased from Sigma-Aldrich (St Louis, MO, USA) unless otherwise indicated.
Plasmid constructs
The FHL2 WT and mutant plasmids were produced by PCR amplification using the following primers: FHL2 WT-F (5′-ACGGAATTCAAATGACTGAGCGCTTTGAC-3′) with FHL2 WT-R (5′-TTACTCGAGTCAGATGTCTTTCCCACAGT-3′); FHL2 WT-F with FHL2 ▵C1-R (5′-CATCTCGAGTTAGTCACAGAAGCAGTTCAGGCA-3′); FHL2 WT-F with FHL2 ▵C2-R (5′-CATCTCGAGTTATTTCTCATAGCAGGGCACACA-3′); FHL2 ▵N1-F (5′-ACGGAATTCAAATGTTCGCCAACACCTGCGAG-3′) with FHL2 WT-R; FHL2 ▵N2-F (5′-ACGGAATTCAAATGGAGTACTCATCCAAGTGC-3′) with FHL2 WT-R; FHL2 ▵N3-F (5′-ACGGAATTCAAATGCAACATGCCATGCAGTGC-3′) with FHL2 WT-R and FHL2 ▵N4-F (5′- GCAGAATTCAAATGAACTGCTTCTGTGACTT-3′) with FHL2 WT-R. PCR products were digested with EcoRI and XhoI (Enzynomics, Seoul, Korea) and ligated into the pCMV-HA or pCMV-Myc empty vectors (Invitrogen, Carlsbad, CA, USA). The pGEX4T1-FHL2 plasmid was cloned using the following primers: FHL2 WT-F (5′-ACGGAATTCATGACTGAGCGCTTTGAC-3′) with FHL2 WT-R. Generation of the pCDNA3 FLAG-tagged or HA-tagged IER3, pCMV-HA Ubiquitin and pCDNA3-MDM2 encoding plasmids were described in previous studies.15, 45, 46 E3 ligase-coding plasmids were generous gifts from Dr In Kyoung Lim (Ajou University; Flag-SKP2) and Dr Kang Yell Choi (Yonsei University; Flag-β-TrCP). The pCDNA4 6xHis-Ub plasmid was a generous gifts from Dr Chin Ha Chung (Seoul National University). Utilizing pCDNA3 HA-IER3 as a template, we generated HA-tagged IER3 mutants plasmids using the following primers: pCDNA3-F (5′-CAAGCTGGCTAGCGTTTAAAC-3′) with IER3 ▵1-R (5′-CCCGCCGGGGAGCATAATCTGGAACATCAT-3′) and IER3 ▵1-F (5′-AGATTATGCTCCCCGGCGGGGCTCCGGTCC-3′) with IER3 WT-R (5′-GCAGAATTCTTAGAAGGCGGCCGGGTGTTG-3′); IER3 WT-F (5′-CTAGGATCCATGTATCCATATGATGTTCCAGATTATGCTTGTCACTCTCGCAGCTGC-3′) with IER3 ▵2-R (5′-AGGCGCTGGGTCCCGGGATGGTGGAGGGGG-3′) and IER3 ▵2-F (5′-CATCCCGGGACCCAGCGCCTCTCGCGGGCA-3′) with IER3 WT-R; IER3 WT-F with IER3 ▵3-R (5′-GTTCCTCGACGCGCCCGGCAGGGGCCGCTG-3′) and IER3 ▵3-F (5′-TGCCGGGCGC GTCGAGGAACCGAACCCAGC-3′) with IER3 WT-R; IER3 WT-F with IER3 ▵4-R (5′-CACCCTCTTCTGGCAGCTGGCGCCGGACCA-3′) and IER3 ▵4-F (5′-CCAGCTGCCAGAAGAGGGTGTGCCGGCGCC-3′) with IER3 WT-R; IER3 WT-F with IER3 ▵5-R (5′-CGAGGACGGGAGCCATCAGGATCTGGCAGA-3′) and IER3 ▵5-F (5′-CCTGATGGCTCCCGTCCTCGAGCCCTTTAA-3′) with IER3 WT-R; IER3 WT-F with IER3 ▵6-R (5′-GCAGAATTCTTAGGACACAGGGGTGGGCGCCA-3′); IER3 WT-F with IER3 K60R-R (5′-TGCGGCTGCGCCGTCGGTGC-3′); and IER3 K60R-F (5′-CGCGGGCACCGACGGCGCAG-3′); IER3 WT-F with IER3 K84R-R (5′-AGAAGCCTTCGGGCTGGGTT-3′) and IER3 K84R-F (5′-AACCCAGCCCGAAGGCTTCT-3′); IER3 PEST-F (5′-GCAGGATCCATGGGACCCCGGCGGGGC-3′) with IER3 PEST-R (5′-GCAGAATTCTTAAGAGGCGCTGGG-3′). All HA-IER3 WT and mutant plasmids were subcloned into the modified pCDNA3.1 empty vector, which contained a GST tag by NcoI and HindIII restriction enzyme sites. The pCDNA3-GST-MDM2 WT and mutant plasmids were produced by PCR amplification using the following primers: MDM2 WT-F (5′- GCTGAATTCATGGTGAGGAGCAGGCAA-3′) with MDM2 WT-R (5′-GCTCTCGAGCTAGGGGAAATAAGTT-3′); MDM2 WT-F with MDM2 ▵C1-R (5′-GCTCTCGAGCTAAGTTGATGGCTGAGA-3′); MDM2 WT-F with MDM2 ▵C2-R (5′-GCTCTCGAGCTACTCATCATCTTCATCT-3′); MDM2 ▵N1-F (5′-GCTGAATTCATGGAGCACAGGAAAATAT-3′) with MDM2 WT-R and MDM2 ▵N2-F (5′-GCTGAATTCATGAGTGAACATTCAGGT-3′) with MDM2 WT-R. PCR products were digested with EcoRI and XhoI and ligated into the modified pCDNA3-GST empty vector, in which a GST tag was introduced between KpnI and BamHI restriction enzyme sites.
Yeast two-hybrid assay
Yeast two-hybrid screening was performed according to a previous study.47
Immunoprecipitation and immunoblot analysis
HeLa cells were transfected with the indicated plasmids as well as with small interference nucleotides (siRNAs) using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Twenty-four hours after transfection, cell lysates were prepared for immunoprecipitation with GST (GE Healthcare Bio-Sciences AB, Uppsala, Sweden) beads or Dynabeads protein G (Invitrogen) according to manufacturer's instructions. After incubation, the samples were boiled and subjected to SDS–polyacrylamide gel electrophoresis for immunoblotting with the respective antibodies. The protein signal on the membranes was detected using a ChemiDoc XRS+ System Imager (Bio-Rad Laboratories, Hercules, CA, USA) and the intensity of each band was quantified using Quantity One software (Bio-Rad Laboratories). The following antibodies were used in this study: mouse anti-FHL2 (11-134; MBL International, Woburn, MA, USA), mouse anti-c-Myc (631206; Clontech), rabbit anti-HA (H6908) and mouse anti-FLAG M2 (F1804; Sigma-Aldrich), mouse anti-ubiquitin (sc-8017), rabbit anti-IER3 (sc-33171), mouse anti-MDM2 (sc-965), mouse anti-HPV18 E6 (sc-365089), mouse anti-HPV18 E7 (sc-365035), mouse anti-p53 (sc-126), rabbit anti-GFP (sc-8334) and rabbit anti-GAPDH (sc-25778; all from Santa Cruz Biotechnology, Santa Cruz, CA, USA), mouse anti-GST (2624) and rabbit anti-FLAG (2638) antibodies (Cell Signaling, Danvers, MA, USA).
Immunofluorescence analysis
The immunofluorescence analysis was performed as previously described.48
RNA interference
Small-interfering RNA (siRNA) target sequences against FHL2, MDM2, IER3, p53, HPV18 E6 and HPV18 E7 were 5′-AACUGCUUCUGUGACUUGU-3′ (FHL2 #1), 5′-CGAAUCUCUCUUUGGCAAG-3′ (FHL2 #1), 5′-GAAGUUAUUAAAGUCUGUU-3′ (MDM2 #2), 5′-CCAGCCAAAAGGCUUCUCUUU-3′ (IER3 #1), 5′-UUCACCUUCGACCCUCUCC-3′ (IER3 #2), 5′-CACUACAACUACAUGUGUA-3′ (p53), 5′-CUAACUAACACUGGGUUAU-3′ (HPV18 E6) and 5′-CCACCAACGUCACACAAUGU-3′ (HPV18 E7). MDM2 siRNA (#1) was purchased from Bioneer (Daejeon, South Korea). The control siRNA sequence used was 5′-CCUACGCCACCAAUUUCGU-3′. The sense and antisense oligonucleotides were annealed in the presence of the annealing buffer (Bioneer).
RNA extraction and real-time PCR
RNA was extracted from transfected HeLa cells using TRIzol reagent (Invitrogen). The extracted RNA was analyzed by real-time PCR as described previously.15 The nucleotide sequences of primers used for real-time PCR (Bioneer) were IER3-F (5′-CAGCCGCAGGGTTCTCTAC-3′), IER3-R (5′-GATCTGGCAGAAGACGATGGT-3′), GAPDH-F (5′-AGGGGCCATCCACAGTCTT-3′) and GAPDH-R (5′- AGCCAAAAGGGTCATCATCTCT-3′).
Ubiquitination assay
The in vivo and in vitro ubiquitination assays were performed as previously described.46, 49
Recombinant protein purification
The pGEX4T1-FHL2 plasmid was expressed in Rosetta 2 (DE3) competent cells, a derivative strain of Escherichia coli strain BL21 (EMD Millipore, Billerica, MA, USA). Cells were cultured in Luria-Bertani medium and selected with ampicillin (100 μg/ml) and chloramphenicol (34 μg/ml). The cells were grown in a shaking incubator at 37 °C until OD600=0.5–0.6 and then supplemented with 100 μM isopropyl β-d-thiogalactoside (Sigma-Aldrich) dissolved in the Luria-Bertani medium for 2 h at 23 °C. Thereafter, the cells were harvested and washed twice with cold PBS, resuspended in PBS containing a protease inhibitor cocktail and lysed by sonication. The separated supernatant from lysates was linked to GST beads and incubated overnight at 4 °C. Beads were washed and the GST-bound proteins were eluted with 10 mM reduced glutathione. GST-IER3 was purified from mammalian cells through transfection of 293T cells with pCDNA3.1 GST-IER3 plasmids. Twenty-four hours after the transfection, the cells were lysed in radio-immunoprecipitation assay buffer containing 1 mM Na3VO4, 10 mM NaF and protease inhibitor cocktail. The supernatant was then separated from 293T cell lysates and incubated with GST beads overnight at 4 °C. Beads were washed and GST-bound proteins were eluted with 10 mM reduced glutathione in 50 mM Tris (pH 8.0). The GST tag from the GST-MDM2 and GST-FHL2 proteins were removed by incubation with 100 U/ml thrombin for 16 h at 4 °C.
Two-step co-immunoprecipitation
Ten dishes of HeLa cells (4–5 × 106 cells/100-mm dish) were lysed with NP-40 lysis buffer and centrifuged. The supernatant was incubated with BS3-crosslinked anti-IER3-Dynabeads protein G overnight at 4 °C. The first immunoprecipitated samples were washed with precooled PBS-T three times, and the IER3-linked protein complex was eluted with 0.1 M glycine buffer (pH 2.0) for 1–2 min and immediately neutralized in 1 M Tris buffer (pH 10.0). The second round of immunoprecipitation was performed by incubation of the first elute with crosslinked anti-MDM2-Dynabeads protein G overnight at 4 °C.
Analysis of cell cycle by flow cytometry
The cell cycle analysis was performed as previously described.50
5′-Bromo-2′-deoxyuridine cell proliferation assay
The 5′-bromo-2′-deoxyuridine cell proliferation assay was performed using the 5′-bromo-2′-deoxyuridine labeling and detection kit III (Roche) according to the manufacturer’s instructions.
Human tissue sampling and TMA construction
The TMA was constructed with duplicate 1-mm cores of archival paraffin material from 20 patients diagnosed with cervical cancer at Daegu Catholic University Medical Center (DCUMC), Daegu, Republic of Korea, between 2011 and 2014 (Supplementary Table 1). The median age of the patients was 52.5 years, with the ages ranging from 31 to 75 years. The control group included duplicate cores of paraffin-embedded samples of normal cervical tissues obtained from 20 patients (median=46.5 years, range=42–69 year), who were diagnosed with other non-cancerous diseases at the same hospital during the same period, 2011 to 2014. All tissues used were reexamined after hematoxylin and eosin staining by pathologists, before TMA construction using 5-μm-thin section of paraffin-embedded cores. The present study was reviewed and approved by the DCUMC Institutional Review Board. Informed consent was received from all participating patients.
Immunohistochemical analysis
Immunohistochemical staining for IER3, FHL2 and MDM2 was performed on the TMA slides. The procedures were carried out using the Bond Polymer Intense Detection System (Leica Microsystems, Mount Waverley, VIC, Australia) according to the manufacturer’s instruction with minor modifications as previously described.51 After image acquisition, the proportion of positive cells was determined by a strong immunostaining intensity of squamous cell carcinoma relative to other morphological regions of the cervix, and the proportion and ISs were determined by a modified Allred scoring method. The PS of less than 1% positive cells was equal to zero, and 1–100% positive cells were scored from 1 to 5 using a 20-percent scale. The IS was scaled from 0 to 3 with the nucleus staining nonspecifically ‘0’, weakly ‘1’, moderately ‘2’ or strongly ‘3’. For the ROC analysis, the proportion and ISs were then summed to produce total scores of 0–8.
ROC curve
Optimal cutoff values for target antigens in squamous cell carcinoma of the cervix were obtained by the ROC analysis in the experimental sets of the TMA assay. A strong relationship of a selected antigen with cervical cancer was validated by the ROC analysis-generated cutoff values in the TMA set.
Statistical analysis
Multiple comparisons of values were performed by the Student–Newman–Keuls test using SAS version 9.2 (SAS Institute, Cary, NC, USA), whereas the Student’s t-test was used for comparisons with control values. The data are presented as the mean±s.e.m. and differences were considered to be significant if P<0.05. For the immunostaining analysis of human cervical tissues, P-values derived from t-tests are two-sided and P<0.05 was considered statistically significant. Statistical analyses were performed using SPSS v. 17.0 (SPSS, Inc., Chicago, IL, USA) and SigmaPlot (Systat Software, San Jose, CA, USA).
References
Ciechanover A . Intracellular protein degradation: from a vague idea thru the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Cell Death Differ 2005; 12: 1178–1190.
Lee MJ, Lee JH, Rubinsztein DC . Tau degradation: the ubiquitin-proteasome system versus the autophagy-lysosome system. Prog Neurobiol 2013; 105: 49–59.
Pickart CM, Eddins MJ . Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta 2004; 1695: 55–72.
Komander D, Rape M . The ubiquitin code. Annu Rev Biochem 2012; 81: 203–229.
Sun Y . E3 ubiquitin ligases as cancer targets and biomarkers. Neoplasia 2006; 8: 645–654.
Haupt Y, Maya R, Kazaz A, Oren M . Mdm2 promotes the rapid degradation of p53. Nature 1997; 387: 296–299.
Kubbutat MH, Jones SN, Vousden KH . Regulation of p53 stability by Mdm2. Nature 1997; 387: 299–303.
Wu MX . Roles of the stress-induced gene IEX-1 in regulation of cell death and oncogenesis. Apoptosis 2003; 8: 11–18.
Arlt A, Schafer H . Role of the immediate early response 3 (IER3) gene in cellular stress response, inflammation and tumorigenesis. Eur J Cell Biol 2011; 90: 545–552.
Arlt A, Schafer H . Role of the immediate early response 3 (IER3) gene in cellular stress response, inflammation and tumorigenesis. Eur J Cell Biol 2010; 90: 545–552.
Arlt A, Grobe O, Sieke A, Kruse ML, Folsch UR, Schmidt WE et al. Expression of the NF-kappa B target gene IEX-1 (p22/PRG1) does not prevent cell death but instead triggers apoptosis in Hela cells. Oncogene 2001; 20: 69–76.
Schilling D, Pittelkow MR, Kumar R . IEX-1, an immediate early gene, increases the rate of apoptosis in keratinocytes. Oncogene 2001; 20: 7992–7997.
Im HJ, Pittelkow MR, Kumar R . Divergent regulation of the growth-promoting gene IEX-1 by the p53 tumor suppressor and Sp1. J Biol Chem 2002; 277: 14612–14621.
Sebens Muerkoster S, Rausch AV, Isberner A, Minkenberg J, Blaszczuk E, Witt M et al. The apoptosis-inducing effect of gastrin on colorectal cancer cells relates to an increased IEX-1 expression mediating NF-kappa B inhibition. Oncogene 2008; 27: 1122–1134.
Jin H, Suh DS, Kim TH, Yeom JH, Lee K, Bae J . IER3 is a crucial mediator of TAp73beta-induced apoptosis in cervical cancer and confers etoposide sensitivity. Sci Rep 2015; 5: 8367.
Pawlikowska P, Leray I, de Laval B, Guihard S, Kumar R, Rosselli F et al. ATM-dependent expression of IEX-1 controls nuclear accumulation of Mcl-1 and the DNA damage response. Cell Death Differ 2010; 17: 1739–1750.
Genini M, Schwalbe P, Scholl FA, Remppis A, Mattei MG, Schafer BW . Subtractive cloning and characterization of DRAL, a novel LIM-domain protein down-regulated in rhabdomyosarcoma. DNA Cell Biol 1997; 16: 433–442.
Chan KK, Tsui SK, Lee SM, Luk SC, Liew CC, Fung KP et al. Molecular cloning and characterization of FHL2, a novel LIM domain protein preferentially expressed in human heart. Gene 1998; 210: 345–350.
Li HY, Kotaka M, Kostin S, Lee SM, Kok LD, Chan KK et al. Translocation of a human focal adhesion LIM-only protein, FHL2, during myofibrillogenesis and identification of LIM2 as the principal determinants of FHL2 focal adhesion localization. Cell Motil Cytoskeleton 2001; 48: 11–23.
Johannessen M, Moller S, Hansen T, Moens U, Van Ghelue M . The multifunctional roles of the four-and-a-half-LIM only protein FHL2. Cell Mol Life Sci 2006; 63: 268–284.
Hayashi H, Nakagami H, Takami Y, Koriyama H, Mori M, Tamai K et al. FHL-2 suppresses VEGF-induced phosphatidylinositol 3-kinase/Akt activation via interaction with sphingosine kinase-1. Arterioscler Thromb Vasc Biol 2009; 29: 909–914.
Kadrmas JL, Beckerle MC . The LIM domain: from the cytoskeleton to the nucleus. Nat Rev Mol Cell Biol 2004; 5: 920–931.
Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM . Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010; 127: 2893–2917.
Munoz N, Castellsague X, de Gonzalez AB, Gissmann L . Chapter 1: HPV in the etiology of human cancer. Vaccine 2006; 24: S3/1–10.
Bernard HU, Burk RD, Chen Z, van Doorslaer K, zur Hausen H, de Villiers EM . Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology 2010; 401: 70–79.
Moody CA, Laimins LA . Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer 2010; 10: 550–560.
Campo-Fernandez B, Morandell D, Santer FR, Zwerschke W, Jansen-Durr P . Identification of the FHL2 transcriptional coactivator as a new functional target of the E7 oncoprotein of human papillomavirus type 16. J Virol 2007; 81: 1027–1032.
Manzo-Merino J, Massimi P, Banks L, Lizano M . High risk HPV E6 oncoproteins impair the subcellular distribution of the four and a half LIM-only protein 2 (FHL2). Virology 2015; 476: 100–105.
Chen J . Signaling pathways in HPV-associated cancers and therapeutic implications. Rev Med Virol 2015; 25 (Suppl 1): 24–53.
Scheffner M, Huibregtse JM, Vierstra RD, Howley PM . The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 1993; 75: 495–505.
Dyson N, Howley PM, Munger K, Harlow E . The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 1989; 243: 934–937.
Heck DV, Yee CL, Howley PM, Munger K . Efficiency of binding the retinoblastoma protein correlates with the transforming capacity of the E7 oncoproteins of the human papillomaviruses. Proc Natl Acad Sci USA 1992; 89: 4442–4446.
Hwang SG, Lee D, Kim J, Seo T, Choe J . Human papillomavirus type 16 E7 binds to E2F1 and activates E2F1-driven transcription in a retinoblastoma protein-independent manner. J Biol Chem 2002; 277: 2923–2930.
Kleiber K, Strebhardt K, Martin BT . The biological relevance of FHL2 in tumour cells and its role as a putative cancer target. Anticancer Res 2007; 27: 55–61.
Zienert E, Eke I, Aust D, Cordes N . LIM-only protein FHL2 critically determines survival and radioresistance of pancreatic cancer cells. Cancer Lett 2015; 364: 17–24.
Ding L, Wang Z, Yan J, Yang X, Liu A, Qiu W et al. Human four-and-a-half LIM family members suppress tumor cell growth through a TGF-beta-like signaling pathway. J Clin Invest 2009; 119: 349–361.
Ng CF, Ng PK, Lui VW, Li J, Chan JY, Fung KP et al. FHL2 exhibits anti-proliferative and anti-apoptotic activities in liver cancer cells. Cancer Lett 2011; 304: 97–106.
Mille F, Thibert C, Fombonne J, Rama N, Guix C, Hayashi H et al. The Patched dependence receptor triggers apoptosis through a DRAL-caspase-9 complex. Nat Cell Biol 2009; 11: 739–746.
Qian Z, Mao L, Fernald AA, Yu H, Luo R, Jiang Y et al. Enhanced expression of FHL2 leads to abnormal myelopoiesis in vivo. Leukemia 2009; 23: 1650–1657.
Wu MX, Ustyugova IV, Han L, Akilov OE . Immediate early response gene X-1, a potential prognostic biomarker in cancers. Expert Opin Ther Targets 2013; 17: 593–606.
Kirkin V, Dikic I . Ubiquitin networks in cancer. Curr Opin Genet Dev 2011; 21: 21–28.
Lipkowitz S, Weissman AM . RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat Rev Cancer 2011; 11: 629–643.
Adams J . The proteasome: a suitable antineoplastic target. Nat Rev Cancer 2004; 4: 349–360.
Balsas P, Galan-Malo P, Marzo I, Naval J . Bortezomib resistance in a myeloma cell line is associated to PSMbeta5 overexpression and polyploidy. Leuk Res 2012; 36: 212–218.
Yoon S, Ha HJ, Kim YH, Won M, Park M, Ko JJ et al. IEX-1-induced cell death requires BIM and is modulated by MCL-1. Biochem Biophys Res Commun 2009; 382: 400–404.
Kim JH, Kim YH, Kim HM, Park HO, Ha NC, Kim TH et al. FOXL2 posttranslational modifications mediated by GSK3beta determine the growth of granulosa cell tumours. Nat Commun 2014; 5: 2936.
Bae J, Donigian JR, Hsueh AJ . Tankyrase 1 interacts with Mcl-1 proteins and inhibits their regulation of apoptosis. J Biol Chem 2003; 278: 5195–5204.
Ryou SM, Yeom JH, Kang HJ, Won M, Kim JS, Lee B et al. Gold nanoparticle-DNA aptamer composites as a universal carrier for in vivo delivery of biologically functional proteins. J Control Release 2014; 196: 287–294.
Jung CR, Lim JH, Choi Y, Kim DG, Kang KJ, Noh SM et al. Enigma negatively regulates p53 through MDM2 and promotes tumor cell survival in mice. J Clin Invest 2010; 120: 4493–4506.
Kim JH, Bae J . Differential apoptotic and proliferative activities of wild-type FOXL2 and blepharophimosis-ptosis-epicanthus inversus syndrome (BPES)-associated mutant FOXL2 proteins. J Reprod Dev 2014; 60: 14–20.
Suh DS, Oh HK, Kim JH, Park S, Shin E, Lee K et al. Identification and validation of differential phosphorylation sites of the nuclear FOXL2 protein as potential novel biomarkers for adult-type granulosa cell tumors. J Proteome Res 2015; 14: 2446–2456.
Acknowledgements
This research was supported by the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning (2014R1A2A2A01006839; NRF-2015R1A5A1008958; NRF-2015R1A2A1A15054332) and by the Next-Generation BioGreen 21 Program (PJ01117703), the Rural Development Administration of the Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Jin, H., Lee, K., Kim, YH. et al. Scaffold protein FHL2 facilitates MDM2-mediated degradation of IER3 to regulate proliferation of cervical cancer cells. Oncogene 35, 5106–5118 (2016). https://doi.org/10.1038/onc.2016.54
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.54
This article is cited by
-
The roles of FHL2 in cancer
Clinical and Experimental Medicine (2023)
-
TAB182 aggravates progression of esophageal squamous cell carcinoma by enhancing β-catenin nuclear translocation through FHL2 dependent manner
Cell Death & Disease (2022)
-
TRAIL promotes hepatocellular carcinoma apoptosis and inhibits proliferation and migration via interacting with IER3
Cancer Cell International (2021)
-
Identification and validation of a six-gene signature associated with glycolysis to predict the prognosis of patients with cervical cancer
BMC Cancer (2020)
-
FOXL2 directs DNA double-strand break repair pathways by differentially interacting with Ku
Nature Communications (2020)