Abstract
The nucleolar factor, digestive organ expansion factor (DEF), has a key role in ribosome biogenesis, functioning in pre-ribosomal RNA (pre-rRNA) processing as a component of the small ribosomal subunit (SSU) processome. Here we show that the peripheral sympathetic nervous system (PSNS) is very underdeveloped in def-deficient zebrafish, and that def haploinsufficiency significantly decreases disease penetrance and tumor growth rate in a MYCN-driven transgenic zebrafish model of neuroblastoma that arises in the PSNS. Consistent with these findings, DEF is highly expressed in human neuroblastoma, and its depletion in human neuroblastoma cell lines induces apoptosis. Interestingly, overexpression of MYCN in zebrafish and in human neuroblastoma cells results in the appearance of intermediate pre-rRNAs species that reflect the processing of pre-rRNAs through Pathway 2, a pathway that processes pre-rRNAs in a different temporal order than the more often used Pathway 1. Our results indicate that DEF and possibly other components of the SSU processome provide a novel site of vulnerability in neuroblastoma cells that could be exploited for targeted therapy.
Similar content being viewed by others
Introduction
The small ribosomal subunit (SSU) processome mediates pre-18 S ribosomal RNA (rRNA) processing.1, 2 This complex contains as many as 72 protein components, some of which have catalytic activities, and thus represent potential targets for inhibition by small molecules.3, 4, 5 Digestive organ expansion factor (DEF) is a component of the SSU processome in the nucleolus and is evolutionarily conserved from yeast (named UTP25) to humans (33% identical and 50% similar at the amino acid level, https://blast.ncbi.nlm.nih.gov/).6, 7, 8 In the zebrafish, we have previously shown that Def is localized to the nucleolus,9 and that its homozygous loss of function confers a phenotype of digestive organ hypoplasia due to decreased cell proliferation indicated by lower levels of BrdU incorporation.10 Hepatic haploinsufficiency of def delays liver regeneration and causes scar formation after partial hepatectomy.11
Neuroblastoma is a solid tumor of the peripheral sympathetic nervous system (PSNS) that accounts for 15% of childhood cancer deaths.12, 13 It derives from the sympathoadrenal lineage of neural crest cells and typically arises in the adrenal medulla or sympathetic ganglia.14, 15 MYCN amplification is present in ~25% of neuroblastomas and is associated with advanced stage disease and a poor prognosis.15, 16 Changes in the nucleolus and in ribosome biogenesis occur in malignant cells because of the key role of the ribosome in supporting cell growth and proliferation.17, 18, 19 In many different types of aggressive tumors, including high-risk neuroblastoma, the cells show enlarged and increased numbers of nucleoli, the site of pre-rRNA processing.20 Indeed, the large nucleolar neuroblastoma subtype has prominent nucleoli and very little discernible cytoplasm and is highly associated with a deteriorating clinical course and MYCN gene amplification.21, 22, 23 Several nucleolar factors, including RNA polymerase I (POLI) and BYSL, are dysregulated in human cancer, and inhibition of RNA POLI has been shown to be an effective therapeutic strategy in certain cancers.24, 25, 26 Haploinsufficiency of RPL24, RPL39 and EIF6, factors involved in the ribosome biogenesis pathway, have been shown to suppress oncogene-induced transformation.27, 28, 29 Intriguingly, the DEF gene in humans is located in a region of chromosome 1q whose gain is associated with an unfavorable outcome in neuroblastoma.30, 31 However, the role of pre-rRNA processing in the initiation and maintenance of childhood neuroblastoma, and thus the possibility that this pathway may constitute a site of vulnerability in neuroblastoma compared with normal cells, remains to be investigated.
The zebrafish is an excellent model system for the study of human diseases, including cancer.32 In particular, we have developed transgenic fish that overexpress MYCN in the PSNS, and then develop neuroblastomas that are histologically indistinguishable from the human tumors. In this model, neural crest cells that migrate into the interrenal gland (IRG), the zebrafish counterpart of the human adrenal medulla, become blocked in differentiation at the neuroblast stage and continue to proliferate due to MYCN overexpression. This provides a faithful animal model with which to investigate normal and aberrant signaling pathways that affect the initiation and maintenance of neuroblastoma.33
In this study, we found that the superior cervical ganglion (SCG) of the PSNS is also very hypoplastic in def-deficient zebrafish, coincident with an accumulation of unprocessed pre-rRNAs and an increased ratio of mature 28 S compared to 18 S rRNA. Loss of function of def in the form of haploinsufficiency significantly decreased the penetrance of MYCN-induced neuroblastoma. Conversely, gain of function of DEF robustly accelerated the onset and increased the penetrance of MYCN-induced neuroblastoma. def haploinsufficiency inhibited the development of MYCN-induced hyperplasia via decreased cell proliferation and increased cell apoptosis, whereas DEF overexpression enhanced MYCN-induced sympathoadrenal precursor hyperplasia through increased cell proliferation. A similar phenotype was observed in human neuroblastoma cell lines, where knockdown of DEF inhibited cell growth and induced apoptosis. Interestingly, overexpression of MYCN alone in zebrafish tumors or due to gene amplification in human tumors caused the induction of pre-rRNA processing through Pathway 2, a pre-rRNA cleavage pathway that is normally used less frequently than Pathway 1 and results in some different pre-rRNA intermediates.34 Our results implicate the pre-rRNA processing machinery as an attractive source of proteins for the development and testing of specific inhibitors that may preferentially target neuroblastoma cells with MYCN amplification, which are often resistant to current therapeutic regimens.
Results
Loss of function of def affects sympathetic neuronal development in zebrafish
Our previous studies showed that def expression is ubiquitous at 2 days post fertilization (dpf) and is enriched in the digestive organs and the brain from 3 to 5 dpf. Although major digestive organs in defhi429/hi429 (designated def−/− in this article) homozygous mutant fish are severely hypoplastic from 3 dpf,10 the effects of loss of def on PSNS development have not previously been evaluated. In yeast, the orthologue of Def is Utp25, a nucleolar protein and component of the SSU processome that is essential for pre-rRNA processing.6, 7 To study the effects of loss of def on PSNS development and the associated consequences in terms of pre-rRNA processing, we assessed embryonic sympathetic neuronal development in the context of loss-of-function def−/− mutant fish.
To investigate sympathetic neuronal development in the def−/− mutant line, we first examined the expression of HuC, sox10 and phox2b markers of neural crest cell development and sympathetic lineage differentiation in zebrafish.35, 36, 37, 38, 39 Whole-mount in situ hybridization showed that the expression of HuC, sox10 and phox2b were identical in homozygous def−/− mutants compared with wild-type siblings at 1 dpf (Figures 1a–f), indicating a lack of detectable abnormalities during the early events of neural crest cell development and sympathetic lineage differentiation in animals with homozygous loss of def. To evaluate differentiation of neural crest cells into the sympathoadrenal lineage, we used riboprobes to the tyrosine hydroxylase (th) and dopamine-β-hydroxylase (dβh) messenger RNAs, which encode two enzymes required for catecholamine synthesis, as markers. Strikingly, def−/− homozygous mutants showed a reduction in sympathetic neuronal cells expressing th and dβh in the SCG at 3 dpf, as compared with wild-type siblings (Figures 1g–l). We also bred def mutant fish with the Tg(dβh:EGFP) transgenic line (TgEGFP),33 and SCG development was evaluated by EGFP intensity at 5 dpf. Although def wild-type and heterozygous mutant embryos showed comparable levels of EGFP intensity in the SCG, sympathetic neuronal cells expressing EGFP were dramatically reduced in def−/− homozygous mutant fish at 5 dpf (Figures 1m–q), indicating that the SCG was severely hypoplastic in the absence of def. The homozygous def mutants then began to die at 10 dpf, with none surviving beyond 14 dpf (data not shown).
Loss of function of def affects sympathetic neuronal development in zebrafish. (a–j) Whole-mount in situ hybridization for HuC (a and b), sox10 (c and d), phox2b (e and f), th (g and h) and dβh (i and j) in def−/− mutant and sibling fish at the indicated genotypes and time points. (k–m) SCG signal intensity indicated by th (g and h), dβh (i and j) or EGFP (n–q) in fish of different def backgrounds was calculated. Horizontal bars indicate the means±s.d., which were compared with the two-tailed unpaired t-test. ***P<0.001; ****P<0.0001. (n–q) Schematic (n) and representative images (o–q) of SCG (arrows) development at 5 dpf in TgEGFP transgenic line with wild-type def (TgEGFP), def+/− (def+/−;TgEGFP) or def−/− (def−/−; TgEGFP) background. DA, diencephalic dopaminergic neurons; dpf, days post fertilization; LC, locus coeruleus; ns, not significant; R, retina; SCG, superior cervical ganglion.
Generation of a new zebrafish transgenic line expressing MYCN in the PSNS
To establish a new zebrafish neuroblastoma model with early tumor onset and high penetrance, we generated a stable transgenic zebrafish line, Tg(dβh:EGFP;dβh:MYCN), designated TgMYCN_TT, that coexpresses both EGFP and human MYCN under control of the zebrafish dβh promoter using a coinjection strategy.40 After coinjection, the two transgenes integrate together in the recipient fish, such that the expression of the EGFP fluorescent protein becomes a useful marker for the expression of MYCN and facilitates visualization of tumor development. The tumor cells in this new zebrafish line showed a more undifferentiated morphology, as compared with those in our previously described Tg (dβh:EGFP-MYCN) (TgMYCN) line that expresses MYCN fused to EGFP33 (Supplementary Figures S1a–h vs S1i–l). Normal cells of the IRG, the zebrafish counterpart of the human adrenal gland, in the control TgEGFP line are predominately chromaffin cells and at 7 weeks post fertilization (wpf) express TH, but not HuC or synaptophysin (Supplementary Figures S1m–o), whereas neuroblastoma cells of TgMYCN_TT line strongly express all three neuroblastoma markers, indicating differentiation arrest at the neuronal progenitor cell stage in tumors (Supplementary Figures S1p–r). The TgMYCN_TT line also expresses higher protein levels of MYCN (Supplementary Figure S1s) than of EGFP-MCYN in the TgMYCN line, which likely contributes to the more rapid onset (7 wpf) and increased tumor penetrance (~70% by 29 wpf) of neuroblastoma in the TgMYCN_TT line (Supplementary Figure S1t).
def haploinsufficiency decreases the penetrance and inhibits the growth of neuroblastoma
We next asked whether the expression levels of DEF are limiting for the initiation and progression of neuroblastoma by testing whether haploinsufficiency for def would delay the onset and decrease the penetrance of MYCN-induced neuroblastoma. The def+/− heterozygous mutant fish were bred with the TgEGFP line for analysis of the IRG development, and with the TgMYCN_TT line for tumor watch. def haploinsufficiency did not affect the IRG development evaluated by EGFP intensity at 3 wpf (Supplementary Figures S1u–w). However, def haploinsufficiency remarkably decreased the penetrance of neuroblastoma induced in the TgMYCN_TT line: ~40% in the def+/−;TgMYCN_TT line compared with ~70% in the control TgMYCN_TT line by 21 wpf (P=0.0154), although tumor onset times were identical (Figures 2a–d). The fish with tumors developed at 7 wpf were then followed through 21 wpf, and the tumor size was estimated by the intensity of the EGFP signal. Intriguingly, the tumors in the def+/−; TgMYCN_TT line were much smaller than the ones in the TgMYCN_TT line (Figure 2e), coincident with def expression levels that were only 27% of those in the def+/+;TgMYCN_TT tumors (Supplementary Figure S1x). Zebrafish mycn expression levels were not significantly changed in the def+/−;TgMYCN_TT compared with the def+/+;TgMYCN_TT tumors (Supplementary Figure S1x), indicating that def haploinsufficiency does not affect mycn expression in zebrafish. Because DEF is a nucleolar factor involved in pre-18 S rRNA processing and ribosome biogenesis, we examined frozen sections of the IRG or tumors of 7 wpf fish stained with an antibody to the nucleolar marker, fibrillarin (Fib). The tumor cells of TgMYCN_TT exhibited an increased cross-sectional area of nucleoli compared with controls of both TgEGFP and def+/−;TgEGFP fish, while the nucleoli increased only half as much in the def+/−;TgMYCN_TT line compared with the TgMYCN_TT line (P<0.0001; Figures 2f–j). Thus, our data indicate that def haploinsufficiency not only decreases the penetrance of neuroblastoma, but also inhibits the growth of tumor cells in MYCN-induced, high-risk neuroblastoma (Figures 2d and e).
def haploinsufficiency decreases the penetrance and inhibits the growth of tumors in the TgMYCN_TT neuroblastoma model. (a–c) In TgMYCN_TT transgenic line with wild-type def (TgMYCN_TT) (b) or def+/− (def+/−;TgMYCN_TT) (c) background, EGFP expressed in the tumor (arrows) arose in the IRG at 21 wpf. TgEGFP transgenic line (a) was used as negative control. Scale bar, 1 mm. (d) Cumulative frequency of neuroblastoma in stable transgenic lines analyzed by Kaplan–Meier method. The difference between tumor onset by 21 wpf for TgMYCN_TT and def+/−;TgMYCN_TT is significant at P=0.0154 by the log-rank test. (e) Tumor size in transgenic fish of different backgrounds (b and c) was calculated. Horizontal bars indicate the means±s.e.m., which were compared with the two-tailed unpaired Welch t-test, P=0.0208. (f–i) Coimmunostaining of EGFP and nucleolar marker Fib of the sagittal sections through IRG of TgEGFP (f) and def+/−;TgEGFP (g), and the tumor sections of TgMYCN_TT (h) and def+/−;TgMYCN_TT (i) transgenic lines at 7 wpf. Nuclei were stained with DAPI. Scale bar, 5 μm. (j) Nucleolar area indicated by nucleolar marker Fib in the sections of (f–i) was calculated. Horizontal bars indicate the means±s.e.m., which were compared with the two-tailed unpaired Welch t-test. wpf, weeks post fertilization.
def haploinsufficiency inhibits neuroblastoma growth by decreasing cell proliferation and increasing apoptotic cell death
The effects of def haploinsufficiency on tumor cell growth raise the critical question of whether loss of function of def adversely affects one or more of the three factors determining overall growth rate in neuroblastoma: cell proliferation, differentiation and apoptosis. Using EdU-labeling to determine the cell proliferation rate of the sympathoadrenal lineage in the IRG region of the TgMYCN_TT line, we found that 7.17% of the EGFP+/dβh+ sympathoadrenal cells showed incorporation of EdU after 2 h of pulse labeling, with very few EdU-positive sympathoadrenal cells identified in control TgEGFP and def+/−;TgEGFP fish at 6 wpf (Figures 3a, d–f). By contrast, the fraction of EdU-positive sympathoadrenal cells was significantly decreased to 4.39% in the def+/−;TgMYCN_TT line (P=0.0248 vs the TgMYCN_TT line; Figures 3a, g–i). Meanwhile, activated caspase-3 staining at 6 wpf revealed a significant increase in the number of apoptotic EGFP+/dβh+ sympathoadrenal cells in the def+/−;TgMYCN_TT line compared with the def+/+;TgMYCN_TT line (P=0.0052; Figures 3b, j–o). The decreased proliferation and ongoing apoptosis of the sympathoadrenal cell lineage was reflected by a trend toward lower overall numbers of sympathoadrenal cells in the IRG of the def+/−;TgMYCN_TT than the def+/+;TgMYCN_TT line (P=0.1108; Figure 3c). When TgMYCN_TT transgenic fish with neuroblastomas were analyzed at 7 weeks of age, the tumor cells with def haploinsufficiency also exhibited increased apoptosis (Supplementary Figures S2a–g). These observations account for the smaller tumors in the def haploinsufficient compared with the wild-type def TgMYCN transgenic lines (Figure 2e).
def haploinsufficiency decreases cell proliferation and increases apoptotic cell death of sympathoadrenal lineage cells overexpressing MYCN in the IRG at 6 wpf. (a–c) Fraction of EdU-positive sympathoadrenal cells (a), numbers of activated caspase-3-positive apoptotic sympathoadrenal cells (b) and of total EGFP+/dβh+ sympathoadrenal cells (c) in the IRG of control TgEGFP and def+/−;TgEGFP, TgMYCN_TT and def+/−;TgMYCN_TT transgenic fish were plotted at 6 wpf. Each symbol represents the value for an individual fish. Horizontal bars represent mean values that were compared with the two-tailed unpaired Welch t-test (a and c) or two-tailed Fisher’s exact test (b). (d–i) Coimmunostaining of EGFP and EdU of the sagittal sections through IRG of EdU-labeled TgMYCN_TT (d–f) and def+/−;TgMYCN_TT (g–i) transgenic lines at 6 wpf. Nuclei were stained with DAPI. Arrowheads indicate the EdU-labeled proliferating sympathoadrenal cells. Dotted lines indicate the head kidney (HK) boundary. Scale bar, 20 μm. (j–o) Coimmunostaining of EGFP and activated caspase-3 of the sagittal sections through IRG of TgMYCN_TT (j–l) and def+/−;TgMYCN_TT (m–o) transgenic lines at 6 wpf. Nuclei were stained with DAPI. Arrowheads indicate the activated caspase-3-positive apoptotic sympathoadrenal cells. Dotted lines indicate the HK boundary. Scale bar, 20 μm.
To determine if the sympathoadrenal cells undergo a p53-dependent nucleolar stress response (NSR)41, 42, 43 in the def+/−;TgMYCN_TT line, we analyzed the p53 protein levels of these cells by immunofluorescence staining. We found that p53 protein was markedly increased at 5 wpf in the nucleoli of sympathoadrenal lineage cells in the IRG region of the def+/−;TgMYCN_TT line, compared with very-low p53 levels in the def+/+;TgMYCN_TT line (Supplementary Figures S2h–k). Western blot analysis also showed the elevated expression of p53 and activated caspase-3 in tumors with def haploinsufficiency from each of the Tg_MYCN_TT and TgMYCN lines at 7 wpf (Supplementary Figures S2l and m), indicating the activation of a p53-dependent NSR. To genetically assess the contribution of the p53-dependent NSR in the def+/−;TgMYCN_TT line, we bred def+/−;p53e7/+ compound heterozygous mutant fish with the p53e7/+;TgMYCN_TT line, and the offspring of this cross were subjected to a tumor watch. Consistent with the results in Figure 2d, def haploinsufficiency significantly decreased the penetrance of neuroblastoma in the TgMYCN_TT line by 15 wpf (P=0.0247 between TgMYCN_TT and def+/−;TgMYCN_TT; Supplementary Figure S2n). However, the tumor penetrance was significantly rescued in the def+/−;p53e7/e7;TgMYCN_TT line (P=0.0001 vs def+/−;TgMYCN_TT; Supplementary Figure S2n). Thus, the def+/−;TgMYCN_TT fish have lower tumor penetrance due to G1-phase arrest and apoptosis presumably due to the NSR, which is rescued by loss of p53.
DEF overexpression accelerates MYCN-induced neuroblastoma
Because of the importance of DEF in sympathetic neuronal development and in pre-rRNA processing,6, 9, 44 we asked whether increased expression of DEF cooperates with MYCN in the molecular pathogenesis of neuroblastoma. Again we employed our coinjection strategy40 to generate stable transgenic zebrafish lines that coexpress mCherry and human DEF in the PSNS under control of the zebrafish dβh promoter. Two independent stable transgenic zebrafish lines were established: Tg(dβh:mCherry;dβh:DEF) # 1 and #2, designated TgDEF#1 and #2.
To examine the effects of DEF overexpression on SCG development, we bred TgDEF with the TgEGFP line. Evaluation of EGFP fluorescence at 5 dpf did not indicate any appreciable changes in SCG development between TgDEF and the control Tg(dβh:mCherry) line (TgmCherry; Supplementary Figures S3a–c). To investigate potential synergism between DEF and MYCN in neuroblastoma pathogenesis, we then bred both the TgDEF#1 and #2 lines with the TgMYCN line, which has a relatively later onset and a lower tumor penetrance compared with the TgMYCN_TT line, thus making it easier to detect alterations that accelerate tumor onset. Fish transgenic for DEF alone (TgDEF#1 and #2) did not develop neuroblastoma (Figures 4a–f). Fish transgenic for MYCN only (TgMYCN) or MYCN in the TgmCherry transgenic background (TgmCherry;MYCN) developed neuroblastoma in the IRG with a relatively long latency (beginning at 15–17 wpf) and a relatively low penetrance of 5–10% by 29 wpf (Figures 4a, b, g–j), in agreement with our original findings.33 By contrast, both TgDEF#1;MYCN and TgDEF#2;MYCN double transgenic lines developed tumors in the IRG region with a much earlier onset (beginning at 5 wpf) and exhibited markedly increased penetrance of ~50% by 29 wpf (P<0.0001; Figures 4a, b, k–n). We also bred the TgDEF#2 fish with the TgMYCN_TT line, and obtained significantly increased penetrance of neuroblastoma in the compound TgDEF#2;MYCN_TT line, compared with the TgMYCN_TT line (72.4% in the compound TgDEF#2;TgMYCN_TT line vs 64.1% in the control TgMYCN_TT line by 21 wpf, P=0.0462; Supplementary Figure S3d). Thus, DEF overexpression significantly potentiated the tumor onset and penetrance of the MYCN-only transgenic line (Figures 4a and b and Supplementary Figure S3d).
DEF accelerates tumor onset and increases the penetrance of MYCN-induced neuroblastoma. (a, b) Cumulative frequency of neuroblastoma in two independent DEF stable transgenic lines by Kaplan–Meier analysis. The difference between tumor onset by 29 wpf for TgDEF#1;MYCN (a) and TgDEF#2;MYCN (b) vs TgmCherry;MYCN transgenic fish is significant at P<0.0001 and P<0.0001 (by the log-rank test), respectively. (c, d) A 15 wpf TgDEF transgenic line without detectable EGFP and mCherry-positive tumor in the IRG. (g, h) A 35 wpf TgMYCN transgenic line with an EGFP-positive tumor (arrow) arising in the IRG. (k, l) A 15 wpf TgDEF;MYCN compound transgenic line with an EGFP and mCherry-positive tumor (arrows) arising in the IRG. Scale bars represent 1 mm for c, d, g, h, k and l. (e, f, i, j, m and n) H&E-stained sagittal sections of TgDEF transgenic line (e and f), TgMYCN transgenic line with neuroblastoma (i and j) and TgDEF;MYCN transgenic line with neuroblastoma (m and n). Boxes in e, i and m indicate the IRG, and are magnified in f, j and n, respectively. Arrowheads indicate the nucleoli. Scale bars represent 1 mm for e, i and m, and 20 μm for f, j and n. (o) Nucleolar area indicated by H&E staining in the tumor sections of TgMYCN (j) and TgDEF;MYCN (n) transgenic fish was calculated. Horizontal bars indicate the means±s.e.m., which were compared with the two-tailed unpaired Welch t-test, P<0.0001. (p–s) Coimmunostaining of EGFP, mCherry and Fib of the sagittal sections through IRG of TgEGFP (p), TgDEF (q), TgMYCN (r) and TgDEF;MYCN (s) transgenic lines at 6 wpf. Nuclei were stained with DAPI. Scale bar, 4 μm. (t) Nucleolar area indicated by staining with the nucleolar marker Fib in the sections of (p–s) and Figure 2h was calculated. Horizontal bars indicate the means±s.e.m., which were compared with the two-tailed unpaired Welch t-test. E, eye; G, gill; H, heart; I, intestine; IRG, interrenal gland; L, liver; T, testis; wpf, weeks post fertilization.
DEF expression was readily detected in both TgDEF#1;MYCN and TgDEF#2;MYCN tumors, compared with the TgMYCN line (Supplementary Figures S3e and f). Hematoxylin and eosin (H&E) staining showed that MYCN-induced tumors in the IRG were composed of typical undifferentiated, small and round tumor cells that appeared very similar in fish with or without DEF overexpression (Figures 4i, j, m and n). We also examined the nucleoli in these different tumor genotypes, observing that the cross-sectional area of the nucleoli in H&E-stained cells is significantly increased in TgDEF;MYCN double transgenic fish compared with TgMYCN-only transgenic fish (P<0.0001; Figures 4j, n and o). To demonstrate the increased nucleolar area more clearly, we also examined frozen sections through the IRG of 6 wpf fish stained with the antibody against Fib. The nucleoli of sympathetic neuronal progenitor cells in the TgDEF line did not show a larger cross-sectional area, compared with the control TgEGFP line (Figures 4p, q and t). Nucleoli in IRG cells of the TgMYCN line were significantly larger than the control TgEGFP line, and interestingly, nucleoli in the TgDEF;MYCN double transgenic fish were even larger than in the TgMYCN line (Figures 4p and r–t), although they were smaller than in the TgMYCN_TT line (Figure 2h and Figures 4s and t). Taken together, these results indicate that the stimulus driving increased nucleolar size is provided by MYCN, which can be potentiated by increased DEF levels.
Normal cells of the IRG at 15 weeks of age express TH, but not HuC or synaptophysin, consistent with their maturation into chromaffin cells (Supplementary Figures S3g–i). MYCN-transformed neuroblastoma cells stained intensively with all three markers in tumors with or without DEF overexpression (Supplementary Figures S3j–o), indicating differentiation arrest at the neuronal progenitor cell stage. Taken together, our results clearly demonstrate synergism between DEF and MYCN in two independent TgDEF transgenic lines, implicating high levels of expression of this nucleolar protein as a cooperating factor in the pathogenesis of neuroblastoma (Figures 4a and b).
DEF overexpression enhances MYCN-induced hyperplasia and increases cell proliferation in the sympathoadrenal lineage
Our previous studies showed that activated anaplastic lymphoma receptor tyrosine kinase provides prosurvival signals that block MYCN-induced developmentally timed apoptosis, thus allowing the progression of sympathoadrenal neuroblast hyperplasia and promoting rapid progression to neuroblastoma in zebrafish.33 Using a similar approach we investigated the mechanism underlying the synergism between MYCN and DEF overexpression in neuroblastoma pathogenesis. In the TgDEF line expressing DEF alone, the numbers of mCherry+/dβh+ sympathoadrenal cells in the sections of the IRG region were similar to the EGFP+/dβh+ sympathoadrenal cells in control TgEGFP transgenic line at both 5 and 6 wpf (Supplementary Figures S4a–f, m and n), indicating that sympathoadrenal cell development is not affected in transgenic lines expressing DEF alone. By contrast, the numbers of EGFP+/mCherry+/dβh+ sympathoadrenal cells were significantly increased in the TgMYCN and TgDEF;MYCN lines, as compared with those in the control TgEGFP and TgDEF lines at 5 wpf (P=0.0197; Supplementary Figure S4m). At 6 wpf, EGFP+/dβh+ sympathoadrenal cell numbers in the TgMYCN-only fish remained relatively constant (Supplementary Figures S4g–i, m and n), whereas the EGFP+/mCherry+/dβh+ sympathoadrenal cells in the TgDEF;MYCN line continued to expand in the IRG region, so that 10 of the 13 fish examined had increased numbers of these cells (Supplementary Figures S4j–l, m and n). Hence, the number of sympathoadrenal cells in the IRG region of TgDEF;MYCN line is significantly higher than that in the TgMYCN-only line at 6 wpf (P=0.0259), and there is an increased fraction of the fish demonstrating hyperplastic sympathoadrenal cells in the IRG region.
To explore the mechanism involved in the expansion of the sympathoadrenal lineage in the IRG of transgenic fish coexpressing DEF and MYCN, we first assessed the level of apoptotic cell death using activated caspase-3 staining. As we observed previously, apoptotic EGFP+/dβh+ sympathoadrenal cells with positive staining for activated caspase-3 were present in the IRG region in 6 of 11 TgMYCN fish, compared with a lack of detectable apoptotic cells in control TgEGFP and TgDEF fish at 5 wpf (Supplementary Figure S4o). Similarly, a subset of TgDEF;MYCN fish also showed activated caspase-3+/EGFP+/mCherry+/dβh+ apoptotic sympathoadrenal cells in the IRG region (P=0.0709 vs the TgMYCN line; Supplementary Figure S4o), indicating that DEF overexpression differs from overexpression of activated anaplastic lymphoma receptor tyrosine kinase,33 in that DEF does not rescue MYCN-induced developmentally timed apoptosis of sympathoadrenal lineage in the IRG region at 5 wpf. However, EdU-labeling showed that EdU-positive EGFP+/dβh+ sympathoadrenal cells were significantly increased to 21.65% in the TgDEF;MYCN line, compared with 7.62% in the TgMYCN line (P=0.0014; Supplementary Figures S4p–v), consistent with the sympathoadrenal hyperplasia observed in this line at 6 wpf (Supplementary Figures S4j–l and n). Thus, DEF overexpression causes increased proliferation of the sympathoadrenal cell lineage, which can overcome the effects of MYCN-induced developmentally timed apoptosis, leading to an earlier onset and higher penetrance of tumors in transgenic fish overexpressing both DEF and MYCN.
Def and MYCN affect 18 S rRNA maturation and ribosome biogenesis in zebrafish
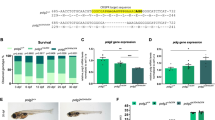
Complete knockout of def, but not def haploinsufficiency, leads to pre-rRNA processing defects in developing zebrafish embryos. Similar to other well-studied species, the mature rRNAs of zebrafish are processed and assembled from an initial pre-rRNA (Figure 5a).45, 46, 47, 48 When independently measured with an Agilent 2100 Bioanalyzer (Santa Clara, CA, USA), the mature 18 S rRNA levels in def−/− homozygous mutants were decreased compared with findings in def+/− heterozygous and wild-type embryos at 5 dpf, whereas 28 S rRNA levels were unaffected (Supplementary Figures S5a–c). Thus, the ratio of 28 S/18 S rRNA was significantly increased in homozygous def mutant embryos, due to a relative paucity of the mature 18 S rRNA (3.3 in def homozygous mutants vs 1.9 and 2.1 in def heterozygous and wild-type fish; Figure 5b). To determine the effect of decreased levels of def on pre-rRNA processing in zebrafish embryos, we analyzed pre-rRNAs from wild-type, def+/− and def−/− zebrafish embryos at 5 dpf by northern blotting. Pre-rRNAs were detected with radioactively labeled oligonucleotide probes complementary to the 5’ external transcribed sequence (5’ETS) and to the internal transcribed spacer 1 (ITS1) (Figure 5a). A probe complementary to the 7SL RNA of the signal recognition particle was used as a loading control.49, 50 def heterozygous fish do not show detectable changes in the levels of pre-rRNAs, indicating the lack of pre-rRNA processing defects in the def+/− embryos (Figures 5c and d). However, def homozygous mutants do show unambiguous accumulation of all the pre-rRNAs detected with the 5’ETS and ITS1 probes, indicating pre-rRNA processing defects in pre-18 S rRNAs when def is not present (pre-rRNAs a, d, and e in Figures 5c and d). Quantitation by ratio analysis of multiple precursors (RAMP)51 revealed two-fold decreases in pre-rRNA e relative to pre-rRNAs a and d (Supplementary Figures S6a and b) and three-fold increases in pre-rRNAs a and d relative to the 7SL loading control for def−/− embryos (Supplementary Figures S6c and d). Thus, only complete loss of def, but not def heterozygosity, leads to defects in pre-18 S rRNA processing in developing zebrafish embryos.
DEF and MYCN regulate pre-rRNA processing in zebrafish. (a) Diagram of pre-rRNA processing in zebrafish cells. The primary pre-rRNA transcript, labeled a, is usually cleaved to form pre-rRNAs d and e through Pathway 1. Pre-rRNAs b and c are formed when Pathway 2 is used. Oligonucleotide probes are indicated (5’ ETS, ITS1). This scheme is based upon previously published findings.45, 46, 47, 48, 76, 77 The presence of Pathway 2 is based upon processing pathways previously described in humans34, 78 and Xenopus.61, 62 (b) Ratios of the mature rRNAs (28 S/18 S) from 5 dpf wild-type (WT), def+/− and def−/− mutant embryos indicated in Supplementary Figures S5a–c were calculated. Data are represented as means±s.e.m. of four single embryos in each group. Mean values were compared with the two-tailed unpaired t-test. **P<0.01. (c, d) Northern blot analysis of pre-rRNA processing in 5 dpf WT, def+/− and def−/− mutant embryos using the 5’ETS (c) or the ITS1 probe (d). A probe for the 7SL RNA was used as a loading control. The pre-rRNA processing intermediates are illustrated to the right. (e, f) Northern blot analysis of pre-rRNA processing in WT, DEF and/or MYCN transgenic zebrafish embryos or zebrafish neuroblastoma samples using the 5’ETS (e) or the ITS1 probe (f). Total RNA was extracted from 5 dpf WT and def−/− zebrafish embryos (lanes 1 and 2) and tumors of the indicated adult transgenic zebrafish lines (lanes 3–10). A probe for the 7SL RNA was used as a loading control. Quantitation of pre-rRNAs on northern blots of c–f by RAMP is shown in Supplementary Figure S6. ns, not significant.
Given the massive effects of MYCN overexpression on transcript levels of ribosome components in neuroblastoma,52 we assessed the impact of this variable on pre-rRNA processing in zebrafish. We first measured the ratio of 28 S/18 S rRNA in the tumors of TgMYCN_TT and def+/−;TgMYCN_TT lines, and observed no significant difference based on def copy number (Supplementary Figure S5d). Using the RNA samples of normal IRG cells sorted from the control TgEGFP fish and MYCN expressing tumor cells (TgMYCN and TgMYCN_TT), we performed quantitative real-time PCR to measure levels of messenger RNAs for five components of the fish SSU processome. The results show increased transcription of def, nob1, and dhx37, but stable levels of transcripts for bms1 and ddx10 (Supplementary Figure S5e), suggesting that the transcription of a subset of the SSU processome components, including def, would appear to be transcriptionally upregulated by MYCN. We also measured the expression of five components of the human SSU processome in MYCN-overexpressing SH-SY5Y_MYCN and SK-N-AS_MYCN stable lines, and the results are largely in agreement with the data that we obtained in zebrafish, with DEF and NOB1 showing the most consistent increase in expression levels when MYCN is overexpressed (Supplementary Figure S5f). Indeed, analysis of the CHIP-seq data from the GEO database (Dr James Bradner, Series GSE80154) showed that high levels of H3K27Ac and MYCN localized to the DEF gene promoter in the neuroblastoma cell lines Kelly, BE2C and NGP, which each has MYCN gene amplification and high levels of MYCN expression (Supplementary Figure S5g). Directly under the modal MYCN binding site, the DEF promoter contains a canonical E-box motif 5′-CACGTG-3′, which is a MYCN binding motif.53 These results show that MYCN binds to the DEF promoter and thereby directly regulates the transcription of DEF. Total RNA was extracted either from 5 dpf zebrafish embryos or tumors dissected from adult fish and analyzed by northern blotting with probes for zebrafish pre-rRNAs or for the 7SL RNA as a loading control. Northern blots with a probe complementary to the 5’ETS (Figure 5a) revealed pre-rRNA species marked b for all samples where MYCN was overexpressed (Figure 5e, lanes 3–10). This was confirmed by quantification using RAMP51 (Supplementary Figures S6e and g). Although not readily detectable in northern blots of zebrafish pre-rRNAs,45, 46, 47, 48 pre-rRNA b likely corresponds to a pre-18S rRNA containing a 5’ extended sequence as it does not hybridize with a probe complementary to ITS1 (Figure 5f). Northern blot analysis with a probe in ITS1 revealed moderate accumulation of pre-rRNA species c relative to the primary transcript for all MYCN-overexpressing tumors (Figure 5f, lanes 3–10; quantitation in Supplementary Figures S6f and h). Taken together, overexpression of MYCN alters pre-rRNA processing in zebrafish tumors, leading to a switch in the frequency of the use of Pathway 2 for the generation of mature rRNAs.
DEF is highly expressed in high-risk human neuroblastomas with MYCN amplification
To determine whether DEF overexpression in zebrafish reflects the expression patterns in human neuroblastoma, we began by analyzing the R2: genomics analysis and visualization platform (http://r2.amc.nl). Using this platform, we analyzed human DEF expression levels in 649 neuroblastomas samples,54 and observed significantly elevated expression of DEF in tumor samples from high-risk patients with tumor cell MYCN gene amplification (Figure 6a). DEF expression levels are also increased in patients with an advanced tumor stage and age >18 months (Figures 6b and c). Low DEF expression is strongly correlated with higher event-free survival probability in both the total cohort and MYCN non-amp subcohort (Figures 6d and e). No significant difference of the event-free survival probability is observed between DEF high and DEF low groups in MYCN-amp subcohort (Figure 6f), because most of the patients in this subcohort already have high MYCN and DEF expression and show unfavorable outcome. Thus, our results indicate elevated levels of DEF expression in MYCN-amplified and other high-risk human neuroblastomas, indicating that the increased DEF levels in MYCN transgenic zebrafish tumors faithfully reproduce what is observed in human neuroblastoma.
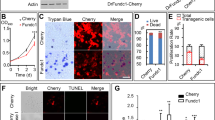
DEF knockdown induces cell growth inhibition and apoptosis in human neuroblastoma cell lines. (a–c) All data used for plotting are from Tumor Neuroblastoma - Kocak - 649 database in R2: genomics analysis and visualization platform (http://r2.amc.nl). The expression of human DEF in neuroblastoma samples (n=649) was analyzed according to MYCN status (MYCN non-amp vs amp) (a), neuroblastoma stages (ST1– 4 S) (b) and age group (c), as indicated. Boxes indicate the median (horizontal line), 25th percentile and 75th percentile; Whiskers, distances from the largest and smallest value to each end of the box that are within 1.5 × box length; Dots, outliers. Mean values were compared with the two-tailed unpaired t-test. ****P<0.0001. (d–f) Kaplan–Meier survival analysis of the total Kocak cohort (d), MYCN non-amp subcohort (e) and MYCN-amp subcohort (f) according to DEF expression. The R2 system gives the cut-off value of DEF expression levels. The difference between the curves for DEF high and DEF low groups are compared with χ2-test. (g–j) Two DEF-targeting shRNAs (shDEF #4 and #7) as well as control luciferase (shLuc) and GFP (shGFP) shRNAs were transduced by lentivirus infection into Kelly (g and h), SH-SY5Y (g and i) and SK-N-AS (g and j) cells. The expression of DEF, p53 and Cleaved PARP (g) were detected by western blot analysis; GAPDH was used as a loading control. Relative cell growth (h–j) at days 2, 4 and 6 after puromycin selection was evaluated. Values are means±s.e.m. of triplicate experiments. (k, l) Analysis of cell apoptosis 3 days post infection by flow cytometric in different phases of the cell cycle after TUNEL and PI staining. Values are means±s.e.m. of triplicate experiments. Mean values from the shDEF#4 and #7 experiments were compared with shGFP by the two-tailed unpaired t-test. *P<0.05, **P<0.01, ***P<0.001. n.s., non-specific band.
DEF knockdown induces cell growth inhibition and apoptosis in human neuroblastoma cell lines
To verify that DEF overexpression is essential for neuroblastoma cell survival, not only in our MYCN transgenic zebrafish models but also in human neuroblastoma cell lines, we performed a DEF short hairpin RNA (shRNA) knockdown experiment. Two independent DEF-specific shRNAs (shDEF#4 and #7) were tested and shown to very efficiently deplete DEF levels in Kelly, SH-SY5Y and SK-N-AS neuroblastoma cell lines, as compared with control shRNAs (shLuc and shGFP) (Figure 6g). DEF depletion also increased p53 and cleaved PARP levels (Figure 6g), and very effectively induced cell death in each of the Kelly and SH-SY5Y cell lines (p53 wild-type), resulting in a loss of >50% of the cells within 2 days (Figures 6h and i). However, in the SK-N-AS cell that harbors mutant p53 with loss of transactivation function,55 DEF depletion didn’t alter cleaved PARP levels (Figure 6g) and the cell death was attenuated (Figure 6j). To assess whether apoptosis is the underlying cause of cell death in neuroblastoma cell lines after depletion of DEF, we stained the cells with TUNEL and PI, and found increased levels of TUNEL-positive cells in each phase of the cell cycle in Kelly and SH-SY5Y cells (Figures 6k and l). Taken together, these data indicate that DEF expression is essential for the survival of human neuroblastoma cells in all phases of the cell cycle, and DEF depletion triggers a p53-dependent NSR.
To further test the hypothesis that MYCN overexpression leads to changes in pre-rRNA processing in neuroblastoma, we analyzed pre-rRNA levels by northern blotting in human neuroblastoma cell lines with or without MYCN amplification. We compared northern blots of pre-rRNAs from Kelly and CHP134 neuroblastoma cell lines (MYCN amplified) with those from SH-SY5Y and SK-N-AS neuroblastoma cell lines (MYCN non-amplified). As in zebrafish, human rRNAs are transcribed as a single pre-RNA that is cleaved to form the mature rRNAs (Figure 7a). Following northern blotting with three probes complementary to the pre-rRNAs (P0, P2, P3), we found that MYCN amplification does not consistently affect pre-rRNA levels (Figures 7b–d; quantitation in Supplementary Figures S7a–h), with one exception: the appearance of the 36 S pre-rRNA species. Interestingly, this precursor was detected in both MYCN-amplified cell lines but not in MYCN non-amplified cells (Figure 7d; compare lanes 1–2 to 3–4). The 36 S pre-rRNA is a precursor in the minor Pathway 2 of pre-rRNA processing in humans (Figure 7a),34 and is analogous to the pre-rRNA species c that we observe in zebrafish tumors when MYCN is overexpressed (Figure 5f). To assess whether the use of Pathway 2 is indeed due to MYCN overexpression, we depleted MYCN in the MYCN-amplified neuroblastoma cell lines, Kelly and CHP134, and confirmed depletion of MYCN by western blot (Figure 7e, compare lanes 2–3 to 1 and lanes 5–6 to 4). The DEF protein levels were decreased upon MYCN depletion (Figure 7e), indicating that high levels of DEF expression by these cell lines depend on high levels of MYCN expression, which is consistent with ChIP-seq data showing that the DEF promoter is a direct target of MYCN (Supplementary Figure S5g). Northern blot analysis with probe P3 revealed a decrease in the levels of the 36 S pre-rRNA relative to the 7SL loading control upon depletion of MYCN (Figure 7f; compare lanes 2–3 to 1 and 5–6 to 4). This is consistent with the hypothesis that MYCN overexpression leads to the use of Pathway 2 for pre-rRNA processing. Quantitation of three biological replicates by RAMP (Supplementary Figures S7i and j) revealed that both shMYNC#1 and shMYCN#5 significantly decreased the 36 S pre-rRNA in Kelly cells. In contrast, only shMYCN#5 significantly decreased the 36 S pre-rRNA in CHP134 cells. Although shMYCN#1 did not significantly decrease the 36 S pre-rRNA in CHP134 cells, the levels of the 36 S pre-rRNA are trending toward a reduction. We conclude that similar to MYCN overexpression in zebrafish, MYCN amplification in human neuroblastoma cell lines results in the upregulation of the otherwise little-used Pathway 2 for pre-rRNA processing.
MYCN amplification increases the frequency with which human neuroblastoma cells use Pathway 2 for pre-rRNA processing. (a) Diagram of pre-rRNA processing in human cells. Oligonucleotide probes P0, P2 and P3 are labeled. Pathway 1 and Pathway 2 for pre-rRNA processing are indicated.34, 78 (b–d) Total RNA was isolated from the indicated human neuroblastoma cells lines with or without MYCN amplification as shown, and hybridized with probes P0 (b), P2 (c) and P3 (d). A probe for the 7SL RNA was used as a loading control. Quantitation of pre-rRNAs on northern blots of (b–d) by RAMP is shown in Supplementary Figures S7a–h. (e, f) Two MYCN-targeting shRNAs (shMYCN #1 and #5) and control GFP shRNA (shGFP) were transduced by lentivirus infection into Kelly and CHP134 cells. The expression levels of MYCN and DEF (e) were detected by western blot analysis; α-tubulin was used as a loading control n.s., non-specific band. Total RNA on a northern blot (f) was hybridized with probe P3, and a probe for the 7SL RNA was used as a loading control. Quantitation of pre-rRNAs on northern blot of f by RAMP is shown in Supplementary Figures S7i and j.
Discussion
The nucleolus is the largest structure in the nucleus of eukaryotic cells, serving as the site of rRNA synthesis and ribosome assembly, and thus is essential for the translation of protein in cells.56 Rapidly proliferating cells require elevated levels of protein synthesis to maintain cell growth, accompanied by increases in the number and size of nucleoli.57 In malignant cells, an enlarged nucleolus reflects increased rates of ribosomal DNA (rDNA) transcription and ribosome biogenesis, and in general a larger nucleolar size in primary tumors correlates with a worse clinical prognosis.58 In this study, we show that the TgDEF;MYCN double transgenic fish develop neuroblastoma with an earlier onset and a higher penetrance than is typical of zebrafish TgMYCN line overexpressing MYCN alone (Figures 4a and b). Tumor cells in these TgDEF;MYCN compound transgenic fish have sympathoadrenal precursor cells with an increased nucleolar size and a higher proliferative fraction than found in TgMYCN single transgenic fish (Figures 4o and t and Supplementary Figure S4v). In childhood neuroblastoma, the Shimada group has identified a subtype characterized by large and prominent nucleoli called 'large nucleolar neuroblastoma',22 which is highly associated with MYCN amplification and a poor prognosis.23 Here we show that these neuroblastomas with MYCN amplification also have high levels of expression of DEF, which is an essential component of the multi-protein complex called the SSU processome and required for processing of the pre-rRNA into the 18 s rRNA.6, 7, 8 Thus, it appears likely that overexpression of DEF is directly linked to enlargement of the nucleolus in TgDEF;MYCN double transgenic fish.
How, then, does DEF overexpression act synergistically with MYCN to promote neuroblastoma? Enlarged nucleoli may contain more rRNA for ribosome assembly to promote protein translation and cell proliferation, which we postulate allows the accelerated development of neuroblastoma. Our previous studies have shown that overexpression of MYCN blocks the differentiation of neuroblast precursors and induces developmentally timed apoptosis in the IRG.33 DEF-induced proliferative increases in this cell lineage appear to overcome the effects of MYCN-induced apoptosis, resulting in an earlier onset and higher penetrance of neuroblastoma in transgenic fish overexpressing both of these genes (Figures 4a and b).
We have found that overexpression of MYCN in neuroblastoma cells from two species (human and zebrafish) results in the increased use of Pathway 2 as a pre-rRNA processing pathway. The use of two pathways for pre-rRNA processing has been previously well-documented in vertebrates. In the late 1960 s and early 1970 s, these pre-rRNA processing pathways were proposed based on the observation that pre-rRNAs from HeLa cells represent different temporal sequences of pre-rRNA cleavage.59, 60 Likewise, in Xenopus, pre-rRNA processing can occur by one of two pathways even within a single cell.61 Disruption of domain II of the U3 snoRNA increased levels of the 36 S pre-rRNA in Xenopus cells that process pre-rRNA with two pathways,62 similar to the increase in zebrafish pre-rRNA labeled 'c' in Figure 5f, and in the human 36 S pre-rRNA that we observed with MYCN overexpression (Figure 7d). In mouse and human cells, the pre-rRNA processing pathway choice is shifted toward the formation of 36 S pre-rRNA after depletion of the exonuclease, Xrn2, and after depletion of factors required for large ribosomal subunit biogenesis, including PES1 and RPL26.34, 51, 63 Interestingly, the use of Pathway 2 for pre-rRNA processing is associated with increased cell growth.51
Our results indicate that MYCN overexpression similarly shifts pre-rRNA processing in zebrafish and human neuroblastoma cells to favor early cleavage of the 5’ end of ITS1 through Pathway 2 (Figure 5f and Figure 7d). The formation of the 36 S pre-rRNA results from cleavage near the 5’ end of ITS1 before cleavage at the 3’ end of ITS1.34, 51, 63 This may occur via the large ribosomal subunit biogenesis factor, PES1, as amplification of MYCN in neuroblastoma results in an increase in PES1 levels.64 An intriguing avenue for future studies would be to decipher the molecular mechanism by which MYCN overexpression in neuroblastoma leads to early cleavage at the 5’ end of ITS1. An additional important avenue for future investigation is whether MYCN has a direct role in pre-rRNA transcription. Although it is well-known that c-MYC has an important, direct role in pre-rRNA transcription,65, 66, 67, 68, 69, 70 it is not yet known whether MYCN has a similar role, though previous studies have detected a 45% increase in total RNA levels after MYCN is expressed.52
def loss of function has consequences for neuroblastoma pathogenesis in that def haploinsufficiency decreases the penetrance and inhibits the growth of MYCN-induced neuroblastoma. The nucleoli in the neuroblastoma cells with def haploinsufficiency are much smaller than in neuroblastoma cells arising in fish with wild-type def, resulting in decreased levels of fully processed 18 S rRNA in the cells (Figure 5b and Supplementary Figures S5a–c), which we postulate acts as a rate-limiting step in tumor initiation and progression. Thus, the lower levels of fully processed rRNA in animals with def haploinsufficiency decrease cell proliferation (Figure 3a) and induce apoptosis in neuroblast precursors (Figure 3b), as well as in fully transformed neuroblastoma cells (Supplementary Figure S2g), leading to compromised tumor growth (Figure 2e).
Given the potency of dysregulated MYC as a major driver of human cancers, it is not surprising that many components of the cellular growth machinery are upregulated in such tumors.71 However, identifying useful therapeutic targets among these components has proved a daunting task. Hannan and co-workers17 have shown that small molecule inhibition of RNA POLI, the enzyme responsible for transcribing the rDNA genes, selectively kills MYC-driven lymphoma cells in vivo.24 Furthermore, Laiho and co-workers72 have identified the small molecule, BMH-21, which binds GC-rich sequences found in rDNA, thus inhibiting RNA POLI transcription and inhibiting melanoma and colorectal carcinoma tumor growth in vivo. Our studies of pre-rRNA processing pathways raise the question of whether other components of the ribosome biogenesis pathway, such as the SSU processome, could serve as therapeutic targets in neuroblastomas overexpressing the MYCN oncogene. We show that MYCN-transformed neuroblastoma cells undergo apoptosis due to a lack of sufficient 18 S rRNA in a def haploinsufficient background (Figure 2e and Supplementary Figures S2a–g), suggesting that this pathway provides a unique and previously unexplored site of vulnerability in these tumor cells. In fact, the SSU processome, in which DEF is a component, contains as many as 72 components, including potentially 'druggable' enzymatic components, such as the endoribonuclease NOB1, the GTPase BMS1 and the RNA helicases DDX10, DDX18, DHX37 and DDX49.3, 5 Like DEF, the messenger RNAs encoding these proteins also show elevated expression levels in high-risk neuroblastomas with an advanced stage and MYCN amplification (R2 database, http://r2.amc.nl). Because these proteins are also likely required at high levels by MYCN-amplified neuroblastomas for maintenance of the malignant phenotype, the genes that encode them may have 'synthetic lethal' properties in MYCN-transformed cells when compared with normal cells. Hence, these enzymes may well be productive targets for the development of small-molecule inhibitors that can kill MYCN-overexpressing neuroblastoma cells at doses that are relatively non-toxic to normal tissues. Our work also shows that neuroblastoma cells overexpressing MYCN activate Pathway 2 for pre-rRNA processing, suggesting dependence on as-yet-undefined nucleolar proteins in this processing pathway. Because Pathway 2 is specifically upregulated in MYCN-transformed cells, it will be important to identify its rate-limiting components, which in fact may represent even better targets for therapeutic development in neuroblastomas with MYCN overexpression.
Materials and methods
Zebrafish lines and maintenance
Zebrafish were raised and maintained according to standard procedures. Zebrafish were all of the AB background strain. All zebrafish studies and maintenance were done in accord with Dana-Farber Cancer Institute IACUC-approved protocol #02-107.
Zebrafish lines Tg(dβh:EGFP), Tg(dβh:mCherry), Tg (dβh:EGFP-MYCN),33 defhi429/hi429 10 and p53e7/e7 73 were described previously, designated TgEGFP, TgmCherry, TgMYCN, def−/− and p53e7/e7 in the text, respectively. To generate zebrafish transgenic line Tg(dβh:mCherry;dβh:DEF), DEF was subcloned under the dβh promoter,33 using the primers 5′-GGG GAC AAG TTT GTA CAA AAA AGC AGG CTT TAC CAT GGG CAA ACG CGG GAG CCG GAG C-3′ and 5′-GGG GAC CAC TTT GTA CAA GAA AGC TGG GTT CAT TTT TCT CCA GTA ATG AAG AG-3′. DNA constructs dβh:mCherry and dβh:DEF were linearized by I-SceI, followed by injection into zebrafish embryos at the one-cell stage. Mosaic F0 transgenic fish with germline integration were identified, and two independent Tg(dβh:mCherry;dβh:DEF) transgenic lines (TgDEF#1 and #2) were established. The primer pair DEF_Fw1665: 5′-CCA GGT GGC CGT GAG GAA TGT-3′ and DEF_Rv2148: 5′-GGT CCA CGT GGC CTC TTC TCC TCT-3′ was used to genotype TgDEF#1 and #2 lines using the genomic DNA extracted from fin clips as template. For the Tg(dβh:EGFP;dβh:MYCN) transgenic line, MYCN was subcloned under the dβh promoter from the pcDNA3.1+ vector (a generous gift from Dr John Power at the Children's Hospital, Boston). DNA constructs dβh:EGFP and dβh:MYCN were linearized by I-SceI, followed by injection into zebrafish embryos at the one-cell stage. A stable Tg(dβh:EGFP;dβh:MYCN) transgenic line (TgMYCN_TT) was identified as described above. The primer pair MYCN_Fw244: 5′-CTG CTT GAG AAC GAG CTG TG-3′ and MYCN_Rv971: 5′-AGG CAT CGT TTG AGG ATC AG-3′ was used to genotype the TgMYCN_TT line.
Tumor watch
Transgenic or mutant fish were observed biweekly for evidence of EGFP/mCherry fluorescence tumors starting at 5 wpf. Fish with tumors were separated, genotyped and killed after anesthesia and characterized by histopathological H&E and molecular analysis. The cumulative frequency of neuroblastoma development was analyzed by the Kaplan–Meier method with the log-rank test used to determine statistical significance. The exact sample size is indicated in each figure and legend. The investigators were not blinded to the experimental group.
Whole-mount RNA in situ hybridization, northern blot analysis and quantitative real-time PCR
Riboprobe labeling and whole-mount in situ hybridization were performed as described previously.44 Constructs for making digoxigenin (DIG)-labeled RNA probes to detect HuC, sox10, phox2b, th and dβh were as previously described.33, 74 Whole-mount in situ hybridization and quantification of the th and dβh signals were performed in a blinded way before genotyping.
Northern blot analysis was performed as previously described.75 Total RNA (2 μg) from zebrafish samples and total RNA (3 μg) from human cells were extracted by the TRIzol reagent (Life Technologies, Carlsbad, CA, USA) and separated by gel electrophoresis on a 1% agarose/1.25% formaldehyde gel and then transferred to a nylon membrane (Hybond-XL, GE Healthcare, Little Chalfont, UK). RNA species were detected by hybridization with radiolabeled oligonucleotide probes. A probe for the 7SL RNA was used as a loading control. Oligonucleotide probes are as follows: 5’ETS probe, 5′-CGA GCA GAG TGG TAG AGG AAG AGA GCT CTT CCT CGC TCA-3′; ITS1 probe, 5′-CAG GTT CAC CGT TTT CCG AG-3′; probe P0, 5′-CCT CTC CAG CGA CAG GTC GCC AGA GGA CAG CGT GTC AGC-3′; probe P2, 5′- CCA CGC AAA CGC GGT CGT CGG CAC CGG TCA CGA CTC GGC A-3′; probe P3, 5′- AAG GGG TCT TTA AAC CTC CGC GCC GGA ACG CGC TAG GTA C-3′; zebrafish 7SL probe, 5′-CTG TAT TAC TCA GGC TGC AC-3′; human 7SL probe 5′-TGC TCC GTT TCC GAC CTG GGC CGG TTC ACC CCT CCT T-3′. Northern blots were quantified with a phosphorimager (Bio-Rad Personal Molecular Imager, Hercules, CA, USA). RAMP was performed as described.51
For quantitative real-time PCR, total RNA was treated with DNase I prior to reverse transcription and purified with RNeasy mini kit (Qiagen, Hilden, Germany). First-strand complimentary DNA was synthesized using the SuperScript III First-Strand Synthesis System (Life Technologies). Quantitative real-time PCR was performed on ViiA 7 Real-Time PCR System (Life Technologies) according to the manufacturer’s instructions. Expression data represented were normalized against elongation factor 1a (ef1a). Primer pairs for amplification of Hu-DEF (DEF_Fw1665: 5′-CCA GGT GGC CGT GAG GAA TGT-3′ and DEF_Rv2148: 5′-GGT CCA CGT GGC CTC TTC TCC TCT-3′), Zf-def (Zf-def_Fw128: 5′-AGA TTT TGC GCT TGC CTG AC-3′ and Zf-def_Rv486: 5′-TGT AAA CTC TCC CTC CAT CTC TGC-3′) and ef1a (ef1a_Fw-61: 5′-TTA CCT GGC AAA GGG GAG CAG C-3′ and ef1a_Rv340: 5′-GCA CAG CAC AGT CAG CCT GAG A-3′) controls were used.
Protein extraction and western blot analysis
Protein was extracted from zebrafish tumors and cell lines using RIPA buffer. Western blotting was performed as described previously.44 The Zf-p53 monoclonal antibody A7-C10 and the Hu-DEF rabbit polyclonal antibody have been described.44 Anti-activated caspase-3 rabbit polyclonal antibody was purchased from BD Biosciences (#559565, San Jose, CA, USA); anti-α-Tubulin mouse monoclonal (B-5-1-2) antibody from Sigma (T6074, St Louis, MO, USA); and anti-GAPDH rabbit monoclonal (EPR6256) antibody from Abcam (#128915, Cambridge, UK).
EdU pulse labeling, cryo-sectioning, paraffin sectioning, immunostaining and fluorescence-activated cell sorting
For EdU pulse labeling, EdU from the Click-iT Alexa Fluor 647 Imaging Kit (Life Technologies #C10340) was diluted to 2.5 mg/ml for retro-orbital injection. Each killed fish examined at 5 wpf was injected with 1 μl EdU solution, whereas 1.5 μl was used for 6 wpf fish. Fish were fixed 2 h post injection for cryo-sectioning; EdU detection was performed according to the manufacturer’s protocol.
Cryo-sectioning and immunofluorescence staining were performed as previously described.44 Antibodies against EGFP (Life Technologies #A6455 and #A11120), Fib (Abcam #ab4566) and activated caspase-3 (BD Biosciences #559565) were used as primary antibodies. Secondary antibodies were conjugated with Alexa 488, 568, 647 (Life Technologies). DAPI (Life Technologies #S36973) was used for nuclear staining. Fluorescent images were taken by a Leica SP5X scanning confocal microscope at the Confocal and Light Microscopy core facility at Dana-Farber Cancer Institute.
Paraffin sectioning, H&E staining and immunohistochemistry with primary antibodies against TH (Pel-Freez #P40101, Rogers, AR, USA), HuC/D (Life Technologies #A-21271) and synaptophysin (Millipore #MAB5258, Billerica, MA, USA) were performed at the DF/HCC Research Pathology Core, and fluorescence-activated cell sorting at DFCI Flow Cytometry Core according to standard protocols.
Cell counts and statistical analysis
To analyze the hyperplasia and apoptosis of the sympathoadrenal cells before tumor onset, all sections from each individual fish were scanned by confocal z-stack. A single representative section containing the largest number of sympathoadrenal cells or activated caspases-3-positive sympathoadrenal cells in the IRG was selected and quantified for each individual fish. All the data were compared with the two-tailed unpaired Welch t-test.
To analyze the proliferative capacity of the sympathoadrenal cells before tumor onset, all sections from each individual fish were scanned by confocal z-stack. A single representative section containing the largest ratio of EdU+ sympathoadrenal cells to the total number of sympathoadrenal cells in the IRG was selected and quantified for each individual fish. The data were analyzed by two-tailed unpaired Welch t-test.
To assess apoptosis in tumors, tumor samples from two different TgMYCN_TT or def+/−;TgMYCN_TT transgenic fish were analyzed. Activated caspases-3-positive tumor cells from five random confocal fields of each sample were quantified. The data were compared with two-tailed Fisher’s exact test.
To calculate the nucleolar size, sections were scanned by confocal z-stack. The largest Fib-staining area of each nucleolus was selected and quantified. The two-tailed unpaired Welch t-test was used for the analysis.
Samples for these experiments were randomly selected. No statistical methods were used for sample size estimation. The investigators were not blinded to the experimental group.
Cell culture and shRNAs knockdown in neuroblastoma cell lines
Kelly, SH-SY5Y and SK-N-AS were stocked in our laboratory and maintained in RPMI 1640 Medium (GIBCO, Gaithersburg, MD, USA) supplemented with 10% fetal bovine serum (Sigma) and 1 × penicillin-streptomycin (GIBCO). Absence of mycoplasma contamination was confirmed by MycoAlert Mycoplasma Detection Kit (Lonza #LT07-418, Portsmouth, NH, USA). All the shRNAs constructs against DEF, MYCN, luciferase and GFP cloned into the lentiviral vector pLKO.1 were obtained from the RNAi Consortium (Broad Institute of MIT and Harvard).
Each control, DEF or MYCN shRNA construct was cotransfected with pMD2.G and psPAX2 into HEK293T cells using FUGENE 6 transfection reagent (Promega, Madison, WI, USA). Cell culture medium containing the lentivirus was filtered by 0.45 μm nitrocellulose membrane and infected with neuroblastoma cells in the presence of 8 μg/ml polybrene. After selecting with puromycin for 36 h, cell growth was measured by CellTiter-Glo Luminescent Cell Viability Assay (Promega), and cell apoptosis in each phase of the cell cycle by APO-BrdU TUNEL Assay Kit (Life Technologies) according to the manufacturer’s instructions. The target sequences of the shRNAs used are as follows. shLuc: 5′-CTT CGA AAT GTC CGT TCG GTT-3′; shGFP: 5′-ACA ACA GCC ACA ACG TCT ATA-3′; shDEF#4: 5′-GCT ACT CAA CAC CCT AAC TAA-3′; shDEF#7: 5′-CCG TAT TTG TGG GCA ATA TTG-3′; shMYCN#1: 5′-ACG TGC CGG AGT TGG TAA AGA-3′; shMYCN#5: 5′-CAT ACC TAA GTA CTG TAA TAA-3′.
References
Bernstein KA, Gallagher JE, Mitchell BM, Granneman S, Baserga SJ . The small-subunit processome is a ribosome assembly intermediate. Eukaryot Cell 2004; 3: 1619–1626.
Dragon F, Gallagher JE, Compagnone-Post PA, Mitchell BM, Porwancher KA, Wehner KA et al. A large nucleolar U3 ribonucleoprotein required for 18 S ribosomal RNA biogenesis. Nature 2002; 417: 967–970.
Phipps KR, Charette J, Baserga SJ . The small subunit processome in ribosome biogenesis-progress and prospects. Wiley Interdiscip Rev RNA 2011; 2: 1–21.
Warner JR . The economics of ribosome biosynthesis in yeast. Trends Biochem Sci 1999; 24: 437–440.
Woolford JL Jr, Baserga SJ . Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 2013; 195: 643–681.
Charette JM, Baserga SJ . The DEAD-box RNA helicase-like Utp25 is an SSU processome component. RNA 2010; 16: 2156–2169.
Goldfeder MB, Oliveira CC . Utp25p, a nucleolar Saccharomyces cerevisiae protein, interacts with U3 snoRNP subunits and affects processing of the 35 S pre-rRNA. FEBS J 2010; 277: 2838–2852.
Harscoet E, Dubreucq B, Palauqui JC, Lepiniec L . NOF1 encodes an Arabidopsis protein involved in the control of rRNA expression. PLoS One 2010; 5: e12829.
Tao T, Shi H, Huang D, Peng J . Def functions as a cell autonomous factor in organogenesis of digestive organs in zebrafish. PLoS One 2013; 8: e58858.
Chen J, Ruan H, Ng SM, Gao C, Soo HM, Wu W et al. Loss of function of def selectively up-regulates Delta113p53 expression to arrest expansion growth of digestive organs in zebrafish. Genes Dev 2005; 19: 2900–2911.
Zhu Z, Chen J, Xiong JW, Peng J . Haploinsufficiency of Def activates p53-dependent TGFbeta signalling and causes scar formation after partial hepatectomy. PLoS One 2014; 9: e96576.
Cheung NK, Dyer MA . Neuroblastoma: developmental biology, cancer genomics and immunotherapy. Nat Rev Cancer 2013; 13: 397–411.
Maris JM, Hogarty MD, Bagatell R, Cohn SL . Neuroblastoma. Lancet 2007; 369: 2106–2120.
Maris JM . Recent advances in neuroblastoma. N Engl J Med 2010; 362: 2202–2211.
Maris JM, Matthay KK . Molecular biology of neuroblastoma. J Clin Oncol 1999; 17: 2264–2279.
Huang M, Weiss WA . Neuroblastoma and MYCN. Cold Spring Harb Perspect Med 2013; 3: a014415.
Bywater MJ, Pearson RB, McArthur GA, Hannan RD . Dysregulation of the basal RNA polymerase transcription apparatus in cancer. Nat Rev Cancer 2013; 13: 299–314.
Montanaro L, Trere D, Derenzini M . Nucleolus, ribosomes, and cancer. Am J Pathol 2008; 173: 301–310.
Montanaro L, Trere D, Derenzini M . The emerging role of RNA polymerase I transcription machinery in human malignancy: a clinical perspective. Onco Targets Ther 2013; 6: 909–916.
Hein N, Hannan KM, George AJ, Sanij E, Hannan RD . The nucleolus: an emerging target for cancer therapy. Trends Mol Med 2013; 19: 643–654.
Kobayashi C, Monforte-Munoz HL, Gerbing RB, Tanaka H, Yeo SY, Huh TL et al. Enlarged and prominent nucleoli may be indicative of MYCN amplification: a study of neuroblastoma (Schwannian stroma-poor), undifferentiated/poorly differentiated subtype with high mitosis-karyorrhexis index. Cancer 2005; 103: 174–180.
Tornoczky T, Semjen D, Shimada H, Ambros IM . Pathology of peripheral neuroblastic tumors: significance of prominent nucleoli in undifferentiated/poorly differentiated neuroblastoma. Pathol Oncol Res 2007; 13: 269–275.
Wang LL, Suganuma R, Ikegaki N, Tang X, Naranjo A, McGrady P et al. Neuroblastoma of undifferentiated subtype, prognostic significance of prominent nucleolar formation, and MYC/MYCN protein expression: a report from the Children's Oncology Group. Cancer 2013; 119: 3718–3726.
Bywater MJ, Poortinga G, Sanij E, Hein N, Peck A, Cullinane C et al. Inhibition of RNA polymerase I as a therapeutic strategy to promote cancer-specific activation of p53. Cancer Cell 2012; 22: 51–65.
Hannan KM, Sanij E, Rothblum LI, Hannan RD, Pearson RB . Dysregulation of RNA polymerase I transcription during disease. Biochim Biophys Acta 2013; 1829: 342–360.
Wang H, Xiao W, Zhou Q, Chen Y, Yang S, Sheng J et al. Bystin-like protein is upregulated in hepatocellular carcinoma and required for nucleologenesis in cancer cell proliferation. Cell Res 2009; 19: 1150–1164.
Barna M, Pusic A, Zollo O, Costa M, Kondrashov N, Rego E et al. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature 2008; 456: 971–975.
Dave B, Granados-Principal S, Zhu R, Benz S, Rabizadeh S, Soon-Shiong P et al. Targeting RPL39 and MLF2 reduces tumor initiation and metastasis in breast cancer by inhibiting nitric oxide synthase signaling. Proc Natl Acad Sci USA 2014; 111: 8838–8843.
Gandin V, Miluzio A, Barbieri AM, Beugnet A, Kiyokawa H, Marchisio PC et al. Eukaryotic initiation factor 6 is rate-limiting in translation, growth and transformation. Nature 2008; 455: 684–688.
George RE, Attiyeh EF, Li S, Moreau LA, Neuberg D, Li C et al. Genome-wide analysis of neuroblastomas using high-density single nucleotide polymorphism arrays. PLoS One 2007; 2: e255.
Pezzolo A, Rossi E, Gimelli S, Parodi F, Negri F, Conte M et al. Presence of 1q gain and absence of 7p gain are new predictors of local or metastatic relapse in localized resectable neuroblastoma. Neuro Oncol 2009; 11: 192–200.
Etchin J, Kanki JP, Look AT . Zebrafish as a model for the study of human cancer. Methods Cell Biol 2011; 105: 309–337.
Zhu S, Lee JS, Guo F, Shin J, Perez-Atayde AR, Kutok JL et al. Activated ALK collaborates with MYCN in neuroblastoma pathogenesis. Cancer Cell 2012; 21: 362–373.
Sloan KE, Mattijssen S, Lebaron S, Tollervey D, Pruijn GJ, Watkins NJ . Both endonucleolytic and exonucleolytic cleavage mediate ITS1 removal during human ribosomal RNA processing. J Cell Biol 2013; 200: 577–588.
An M, Luo R, Henion PD . Differentiation and maturation of zebrafish dorsal root and sympathetic ganglion neurons. J Comp Neurol 2002; 446: 267–275.
Dutton KA, Pauliny A, Lopes SS, Elworthy S, Carney TJ, Rauch J et al. Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates. Development 2001; 128: 4113–4125.
Kim CH, Ueshima E, Muraoka O, Tanaka H, Yeo SY, Huh TL et al. Zebrafish elav/HuC homologue as a very early neuronal marker. Neurosci Lett 1996; 216: 109–112.
Pei D, Luther W, Wang W, Paw BH, Stewart RA, George RE . Distinct neuroblastoma-associated alterations of PHOX2B impair sympathetic neuronal differentiation in zebrafish models. PLoS Genet 2013; 9: e1003533.
Stanke M, Junghans D, Geissen M, Goridis C, Ernsberger U, Rohrer H . The Phox2 homeodomain proteins are sufficient to promote the development of sympathetic neurons. Development 1999; 126: 4087–4094.
Langenau DM, Keefe MD, Storer NY, Jette CA, Smith AC, Ceol CJ et al. Co-injection strategies to modify radiation sensitivity and tumor initiation in transgenic Zebrafish. Oncogene 2008; 27: 4242–4248.
Dai MS, Lu H . Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J Biol Chem 2004; 279: 44475–44482.
Fumagalli S, Ivanenkov VV, Teng T, Thomas G . Suprainduction of p53 by disruption of 40 S and 60 S ribosome biogenesis leads to the activation of a novel G2/M checkpoint. Genes Dev 2012; 26: 1028–1040.
Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH . Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell 2003; 3: 577–587.
Tao T, Shi H, Guan Y, Huang D, Chen Y, Lane DP et al. Def defines a conserved nucleolar pathway that leads p53 to proteasome-independent degradation. Cell Res 2013; 23: 620–634.
Azuma M, Toyama R, Laver E, Dawid IB . Perturbation of rRNA synthesis in the bap28 mutation leads to apoptosis mediated by p53 in the zebrafish central nervous system. J Biol Chem 2006; 281: 13309–13316.
Boglev Y, Badrock AP, Trotter AJ, Du Q, Richardson EJ, Parslow AC et al. Autophagy induction is a Tor- and Tp53-independent cell survival response in a zebrafish model of disrupted ribosome biogenesis. PLoS Genet 2013; 9: e1003279.
Wang Y, Luo Y, Hong Y, Peng J, Lo L . Ribosome biogenesis factor Bms1-like is essential for liver development in zebrafish. J Genet Genomics 2012; 39: 451–462.
Zhao C, Andreeva V, Gibert Y, LaBonty M, Lattanzi V, Prabhudesai S et al. Tissue specific roles for the ribosome biogenesis factor Wdr43 in zebrafish development. PLoS Genet 2014; 10: e1004074.
Griffin JN, Sondalle SB, Del VF, Baserga SJ, Khokha MK . The ribosome biogenesis factor Nol11 is required for optimal rDNA transcription and craniofacial development in Xenopus. PLoS Genet 2015; 11: e1005018.
Tafforeau L, Zorbas C, Langhendries JL, Mullineux ST, Stamatopoulou V, Mullier R et al. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre-rRNA processing factors. Mol Cell 2013; 51: 539–551.
Wang M, Anikin L, Pestov DG . Two orthogonal cleavages separate subunit RNAs in mouse ribosome biogenesis. Nucleic Acids Res 2014; 42: 11180–11191.
Boon K, Caron HN, van AR, Valentijn L, Hermus MC, van SP et al. N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. EMBO J 2001; 20: 1383–1393.
Murphy DM, Buckley PG, Bryan K, Das S, Alcock L, Foley NH et al. Global MYCN transcription factor binding analysis in neuroblastoma reveals association with distinct E-box motifs and regions of DNA hypermethylation. PLoS One 2009; 4: e8154.
Kocak H, Ackermann S, Hero B, Kahlert Y, Oberthuer A, Juraeva D et al. Hox-C9 activates the intrinsic pathway of apoptosis and is associated with spontaneous regression in neuroblastoma. Cell Death Dis 2013; 4: e586.
Nakamura Y, Ozaki T, Niizuma H, Ohira M, Kamijo T, Nakagawara A . Functional characterization of a new p53 mutant generated by homozygous deletion in a neuroblastoma cell line. Biochem Biophys Res Commun 2007; 354: 892–898.
Boisvert FM, van KS, Navascues J, Lamond AI . The multifunctional nucleolus. Nat Rev Mol Cell Biol 2007; 8: 574–585.
Quin JE, Devlin JR, Cameron D, Hannan KM, Pearson RB, Hannan RD . Targeting the nucleolus for cancer intervention. Biochim Biophys Acta 2014; 1842: 802–816.
Derenzini M, Montanaro L, Trere D . What the nucleolus says to a tumour pathologist. Histopathology 2009; 54: 753–762.
Weinberg RA, Loening U, Willems M, Penman S . Acrylamide gel electrophoresis of HeLa cell nucleolar RNA. Proc Natl Acad Sci USA 1967; 58: 1088–1095.
Weinberg RA, Penman S . Processing of 45 s nucleolar RNA. J Mol Biol 1970; 47: 169–178.
Savino R, Gerbi SA . In vivo disruption of Xenopus U3 snRNA affects ribosomal RNA processing. EMBO J 1990; 9: 2299–2308.
Borovjagin AV, Gerbi SA . U3 small nucleolar RNA is essential for cleavage at sites 1, 2 and 3 in pre-rRNA and determines which rRNA processing pathway is taken in Xenopus oocytes. J Mol Biol 1999; 286: 1347–1363.
Preti M, O'Donohue MF, Montel-Lehry N, Bortolin-Cavaille ML, Choesmel V, Gleizes PE . Gradual processing of the ITS1 from the nucleolus to the cytoplasm during synthesis of the human 18 S rRNA. Nucleic Acids Res 2013; 41: 4709–4723.
Nakaguro M, Kiyonari S, Kishida S, Cao D, Murakami-Tonami Y, Ichikawa H et al. Nucleolar protein PES1 is a marker of neuroblastoma outcome and is associated with neuroblastoma differentiation. Cancer Sci 2015; 106: 237–243.
Chan JC, Hannan KM, Riddell K, Ng PY, Peck A, Lee RS et al. AKT promotes rRNA synthesis and cooperates with c-MYC to stimulate ribosome biogenesis in cancer. Sci Signal 2011; 4: ra56.
Grewal SS, Li L, Orian A, Eisenman RN, Edgar BA . Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat Cell Biol 2005; 7: 295–302.
Kang J, Kusnadi EP, Ogden AJ, Hicks RJ, Bammert L, Kutay U et al. Amino acid-dependent signaling via S6K1 and MYC is essential for regulation of rDNA transcription. Oncotarget 2016; 7: 48887–48904.
Mitchell NC, Tchoubrieva EB, Chahal A, Woods S, Lee A, Lin JI et al. S6 Kinase is essential for MYC-dependent rDNA transcription in Drosophila. Cell Signal 2015; 27: 2045–2053.
Poortinga G, Hannan KM, Snelling H, Walkley CR, Jenkins A, Sharkey K et al. MAD1 and c-MYC regulate UBF and rDNA transcription during granulocyte differentiation. EMBO J 2004; 23: 3325–3335.
Poortinga G, Wall M, Sanij E, Siwicki K, Ellul J, Brown D et al. c-MYC coordinately regulates ribosomal gene chromatin remodeling and Pol I availability during granulocyte differentiation. Nucleic Acids Res 2011; 39: 3267–3281.
Poortinga G, Quinn LM, Hannan RD . Targeting RNA polymerase I to treat MYC-driven cancer. Oncogene 2015; 34: 403–412.
Peltonen K, Colis L, Liu H, Trivedi R, Moubarek MS, Moore HM et al. A targeting modality for destruction of RNA polymerase I that possesses anticancer activity. Cancer Cell 2014; 25: 77–90.
Berghmans S, Murphey RD, Wienholds E, Neuberg D, Kutok JL, Fletcher CD et al. tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc Natl Acad Sci USA 2005; 102: 407–412.
Stewart RA, Arduini BL, Berghmans S, George RE, Kanki JP, Henion PD et al. Zebrafish foxd3 is selectively required for neural crest specification, migration and survival. Dev Biol 2006; 292: 174–188.
Pestov DG, Lapik YR, Lau LF . Assays for ribosomal RNA processing and ribosome assembly. Curr Protoc Cell Biol 2008; Chapter 22: Unit 22.11.
Essers PB, Pereboom TC, Goos YJ, Paridaen JT, Macinnes AW . A comparative study of nucleostemin family members in zebrafish reveals specific roles in ribosome biogenesis. Dev Biol 2014; 385: 304–315.
Wilkins BJ, Lorent K, Matthews RP, Pack M . p53-mediated biliary defects caused by knockdown of cirh1a, the zebrafish homolog of the gene responsible for North American Indian Childhood Cirrhosis. PLoS One 2013; 8: e77670.
Henras AK, Plisson-Chastang C, O'Donohue MF, Chakraborty A, Gleizes PE . An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip Rev RNA 2015; 6: 225–242.
Acknowledgements
We thank John R Gilbert for editorial review of the manuscript and critical comments, and Hillary Layden and Shawna Chamberlin for their excellent care of the zebrafish. This work was supported by an Innovation Award from Alex’s Lemonade Stand Foundation (ATL), NIH grant CA180692 (ATL), NIH grant GM115710 (SJB), a National Natural Science Foundation of China (No. 31330050; JP), a Young Investigator Award from Alex's Lemonade Stand Foundation (TT), NIH grants GM07205, HD007149 and F30DK109582 (SBS), and David Abraham Fellowship (HS).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Tao, T., Sondalle, S., Shi, H. et al. The pre-rRNA processing factor DEF is rate limiting for the pathogenesis of MYCN-driven neuroblastoma. Oncogene 36, 3852–3867 (2017). https://doi.org/10.1038/onc.2016.527
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.527
This article is cited by
-
The myth and the reality of energy recovery from municipal solid waste
Energy, Sustainability and Society (2018)
-
Zebrafish as a model to study neuroblastoma development
Cell and Tissue Research (2018)