Abstract
Melanoma is the most lethal form of skin cancer and treatment of metastatic melanoma remains challenging. BRAF/MEK inhibitors show only temporary benefit due the occurrence of resistance and immunotherapy is effective only in a subset of patients. To improve patient survival, there is a need to better understand molecular mechanisms that drive melanoma growth and operate downstream of the mitogen activated protein kinase (MAPK) signaling. The Krüppel-like factor 4 (KLF4) is a zinc-finger transcription factor that plays a critical role in embryonic development, stemness and cancer, where it can act either as oncogene or tumor suppressor. KLF4 is highly expressed in post-mitotic epidermal cells, but its role in melanoma remains unknown. Here, we address the function of KLF4 in melanoma and its interaction with the MAPK signaling pathway. We find that KLF4 is highly expressed in a subset of human melanomas. Ectopic expression of KLF4 enhances melanoma cell growth by decreasing apoptosis. Conversely, knock-down of KLF4 reduces melanoma cell proliferation and induces cell death. In addition, depletion of KLF4 reduces melanoma xenograft growth in vivo. We find that the RAS/RAF/MEK/ERK signaling positively modulates KLF4 expression through the transcription factor E2F1, which directly binds to KLF4 promoter. Overall, our data demonstrate the pro-tumorigenic role of KLF4 in melanoma and uncover a novel ERK1/2-E2F1-KLF4 axis. These findings identify KLF4 as a possible new molecular target for designing novel therapeutic treatments to control melanoma growth.
Similar content being viewed by others
Introduction
Cutaneous melanoma is the most lethal skin cancer. Whereas patients with early stage disease can often be cured by surgical excision, survival rates in the metastatic stage are poor.1 Over 75% of cutaneous melanomas harbor mutually exclusive activating mutations in BRAF and NRAS, that lead to the constitutive activation of the RAS/RAF/MEK/ERK signaling pathway.2, 3, 4 In the last few years, targeted therapies against BRAF and MEK, and immune checkpoint inhibitors have improved survival of this disease.5 However, BRAF/MEK inhibitors show only temporary benefit due the occurrence of resistance and immunotherapy is effective only in a subset of patients. Thus, to improve patient survival there is a need to better understand molecular mechanisms that drive melanoma growth and operate downstream of MEK/ERK.
We have previously shown that a subset of melanomas and melanoma stem-like cells harbor active Hedgehog signaling and exhibit aberrant expression of stemness genes, including SOX2 and the Krüppel-like factor 4 (KLF4).6, 7, 8 KLF4 was initially identified as a zinc finger transcription factor enriched primarily in post-mitotic, terminally differentiated epithelial cells in the skin and intestine.9, 10 KLF4 is one of the four factors required for reprogramming of adult fibroblasts11 and skin melanocytes12 into induced pluripotent stem cells. KLF4 has also an essential role in the maintenance of genomic stability by modulating DNA damage response and repair processes.13
KLF4 expression and activity are altered in human cancers, although genetic alterations of this gene in cancer are uncommon. KLF4 has a dual role, acting both as a tumor suppressor or oncogene, depending on tissue, tumor type or tumor stage.14 These context-dependent functions appear to be mediated by molecular switches such as p21 and p53,15 through alternative splicing,16 or by post-translational modifications.17, 18 Experimental and clinical evidence has demonstrated the tumor-suppressor role of KLF4 in several types of cancer, including gastric, lung, renal cancers and leukemia,19, 20, 21, 22, 23 and targeted activation of KLF4 has been approved as therapeutic approach of advanced solid tumors.24 Conversely, KLF4 has an oncogenic role in osteosarcoma25 and human breast cancer,26 although in a mouse model it inhibits breast cancer progression and metastasis.27 In pancreatic ductal adenocarcinoma (PDA) KLF4 has a stage-dependent function; it has a pro-tumorigenic function in PDA initiation28 and a tumor-suppressive function in advanced PDA.29 The role of KLF4 in squamous cell carcinoma is controversial.30, 31 The function and regulation of KLF4 in melanoma remains unknown.
Here, we have thoroughly investigated the function of KLF4 in human melanomas and its interaction with the mitogen activated protein kinase (MAPK) signaling. By examining the impact of KLF4 modulation in patient-derived melanoma cells, we provide evidence of the oncogenic role of KLF4 in human melanoma. In addition, we find that the MAPK signaling positively regulates KLF4 through the transcription factor E2F1.
Results
Human melanomas express full length (FL) KLF4 and the isoform KLF4α
To begin to investigate the role of KLF4 in melanoma, we analyzed the expression of KLF4 mRNA in a panel of 28 melanoma tissues. Quantitative real-time PCR (qPCR) analysis revealed that approximately half of the melanomas showed levels of KLF4 mRNA higher than in normal human epidermal melanocytes (Figure 1a). The expression of KLF4 was further analyzed in patient-derived and commercial melanoma cell lines both at the protein and mRNA levels. All samples showed variable expression of KLF4 protein and mRNA (Figure 1b; Supplementary Figure S1a). Western blot and immunofluorescence analyses revealed that endogenous KLF4 was localized mainly in the nucleus (Figures 1c and d).
KLF4 expression in human melanoma. (a) qPCR analysis of full length KLF4 in a panel of 28 human melanomas and normal human epidermal melanocytes (NHEM). The y axis represents expression ratio of gene/(GAPDH and β-ACTIN average). (b) Western blotting analysis of KLF4 protein in a panel of three patients derived melanoma cells (M51, M26c and M33x) and five commercial cell lines (A375, SK-Mel2, SK-Mel 5, SK-Mel28 and Mewo). Actin was used as loading control. (c) Subcellular localization of endogenous KLF4 in A375 melanoma cells. Cell fractionation was performed and lysates were subjected to western blotting with anti-KLF4, anti-GAPDH (control for cytoplasmic proteins) and anti-Fibrillarin (control for nuclear proteins) antibodies. (d) Representative images of A375 cells after immuno-labeling with anti-KLF4 antibody analyzed by fluorescence microscopy. Nuclei were counterstained with 4’,6-diamidino-2-phenylindole. (e) PCR analysis in a panel of four patients derived melanoma cells and five commercial cell lines showing two major KLF4 splicing variants: KLF4 full length (KLF4 FL, 1470 bp) and KLF4α (500 bp). (f) Subcellular localization of transiently transfected KLF4 FL and Myc-tagged KLF4α in M26c melanoma cells. Cell fractionation was performed 48 h after transfection and lysates were subjected to western blotting with anti-KLF4 (for KLF4 FL), anti-Myc (for Myc-KLF4α), anti-GAPDH (control for cytoplasmic proteins) and anti-Fibrillarin (control for nuclear proteins) antibodies. (g) Representative images of KLF4α and KLF4 FL subcellular localization after transient transfection of Myc-tagged KLF4α and KLF4 FL in M26c cells. Immunolocalization was with anti-Myc antibody for Myc-tagged KLF4α (red) and anti-KLF4 for KLF4 FL (green). Nuclei were counterstained with 4’,6-diamidino-2-phenylindole. Scale bar=10 μm.
KLF4 has been shown to express different splicing variants.16, 32 By using the forward and reverse primers that cover both 5' and 3' untranslated regions of KLF4 transcript, we identified the presence of two major bands in most of the tested melanoma cell lines (Figure 1e). Sequence analysis of the complementary DNA (cDNA) clones confirmed that the primary and largest band (1440 bp) was FL KLF4 and the band around 470 bp was isoform α.16 The splice variant KLF4α encodes a truncated protein with deletion of exon 3 that produces a frame shift. As a consequence, KLF4α has the same 33 amino-acid residues at the N-terminal as KLF4 FL, but the rest of amino-acid sequence is different, and lacks the nuclear localization signal and DNA-binding domain (Supplementary Figure S2). To determine the subcellular localization of the two isoforms, M26c cells were transfected with Myc-tagged KLF4α or untagged KLF4 FL. Western blot and immunofluorescence analyses showed that Myc-tagged KLF4α was localized both in the cytoplasm and the nucleus, with a prevalent cytoplasmic distribution. On the contrary, exogenous KLF4 FL was mainly nuclear (Figures 1f and g), consistent with endogenous KLF4 (Figures 1c and d). Melanoma cells displayed variable expression of KLF4α mRNA by qPCR using primers for the specific detection of the isoform α (Supplementary Figure S1b). The closely related KLF family member KLF5 has been shown to cooperate with KLF4 in promoting tumorigenesis and in mediating lapatinib resistance in breast cancer.33 Therefore, we examined the expression of KLF5 in melanoma cells. Interestingly, we found a positive correlation between the expression of KLF4 and KLF5 mRNA in melanoma cells (Supplementary Figure S1c and d).
KLF4 expression is modulated by the RAS/RAF/MEK/ERK signaling in melanoma cells
KLF4 is modulated by a variety of environmental signals, including DNA damage, inflammation and oxidative stress.34 Melanoma is commonly characterized by activation of the RAS/RAF/MEK/ERK pathway, frequently due to mutually exclusive mutations in BRAF and NRAS or other genetic events,35, 36, 37 which leads to constitutive activation of ERK1/2. Consistently, the majority of our melanoma cells showed ERK1/2 phosphorylation in basal conditions (Supplementary Figure S3a). A previous study showed that ERK1 and ERK2 directly phosphorylate KLF4 at Ser123, with consequent KLF4 ubiquitination and degradation, and induction of embryonic stem cell differentiation.18 In addition, Klf4 expression is induced by mutant KRasG12D in a mouse pancreatic acinar cell line.28 Therefore, we sought to investigate whether the RAS/RAF/MEK/ERK signaling might modulate KLF4 in melanoma. Treatment with CI-1040 (PD 184352, 1 μM), a specific MEK1/2 inhibitor,38, 39 drastically reduced KLF4 protein levels in A375, SK-Mel-5 and SK-Mel-28 melanoma cells, harboring BRAF-V600E mutation, and in SK-Mel-2 cells, that carry NRAS-Q61R mutation (Figure 2a). Similar results were obtained using the ERK1/2 inhibitor SCH77298440 (Figure 2b). Consistently, overexpression of BRAF-V600E and BRAF wt in patients-derived M26c melanoma cells and in HEK-293T cells, which harbor wild-type BRAF and NRAS, led to a strong increase in KLF4 protein level (Figure 2c; Supplementary Figure S3b). To further clarify whether regulation of KLF4 by ERK1/2 was exerted at the transcriptional level, qPCR analysis of KLF4 mRNA was performed after treatment of several melanoma cell lines (A375, SK-Mel-2, SK-Mel-5 and SK-Mel-28) with CI-1040 or SCH772984. Expression of KLF4 mRNA was decreased upon treatment with CI-1040 or SCH772984, thus suggesting a transcriptional regulation of KLF4 by the RAS/RAF/MEK/ERK signaling (Figure 2d; Supplementary Figure S3c). The expression of KLF5 was not affected by inhibition of MEK1/2 nor ERK1/2 (Supplementary Figure S3d). The efficiency of both inhibitors was confirmed by disappearance of phosphorylated ERK1/2 (Figures 2a and b) and drastic downregulation of Cyclin D1 (Figure 2e; Supplementary Figure S3c), an established mitogenic target of RAS.41, 42 These results altogether indicate that RAS/RAF/MEK/ERK signaling positively modulates the expression of KLF4 both at the protein and mRNA levels.
RAS/RAF/MEK/ERK signaling induces KLF4 expression in melanoma cells. (a, b) Western blotting analysis showing KLF4 and pERK1/2 expression levels in A375, SK-Mel-2, SK-Mel-5 and SK-Mel-28 melanoma cells treated with MEK1/2 inhibitor CI-1040 (1 μM) (a) or ERK1/2 inhibitor SCH772984 (0.5 μM) (b) for 16 h. Actin was used as loading control. Quantification of KLF4 protein, expressed as relative ratio of KLF4/Actin, is shown in blue. (c) Western blotting analysis of KLF4, BRAF, pERK1/2 in M26c and HEK-293T cells transiently transfected with pBRAF-V600E. Actin was used as loading control. Quantification of KLF4 protein, expressed as relative ratio of KLF4/Actin, is shown in blue. (d, e) Expression of KLF4 and CyclinD1 mRNA in A375, SK-Mel-2, SK-Mel-5 and SK-Mel-28 measured by qPCR after treatment with CI-1040 or SCH772984. The y axis represents expression ratio of gene/(GAPDH and β-ACTIN average), with the level of control equated to 1. Data represent mean±s.e.m. of three independent experiments. *P⩽0.05 and **P<0.01.
RAS/RAF/MEK/ERK signaling modulates the expression of KLF4 through the transcription factor E2F1
Interestingly, we found that in melanoma samples the expression of KLF4 positively correlated with that of the transcription factor E2F1 (P=0.014; Figure 3a), a key transcriptional regulator of proliferation that has a pivotal role in regulating growth and survival in several types of cancer, including melanoma.43, 44 This finding led us to hypothesize that the modulation of KLF4 by the RAS/RAF/MEK/ERK signaling might be mediated by E2F1. To begin to investigate this hypothesis, we tested the effect of MEK1/2 inhibition on E2F1 expression. Western blot analysis showed that CI-1040 treatment almost abolished E2F1 protein levels in four different melanoma cell lines (Figure 3b), indicating that RAS/RAF/MEK/ERK signaling modulates E2F1 expression.
E2F1 mediates the effect of the MAPK signaling on KLF4. (a) Linear correlation analysis of KLF4 with E2F1 expression measured by qPCR in a panel of 28 melanoma tissues (gray dots) and normal human epidermal melanocytes (NHEM) (black dot). Axes represent expression ratio of gene/(GAPDH and β-ACTIN average). The extent of the correlation is indicated by R coefficient. (b) Western blotting analysis showing E2F1 and pERK1/2 expression levels in A375, SK-Mel-2, SK-Mel-5 and SK-Mel-28 cells treated with CI-1040 (1 μM) for 16 h. Actin was used as loading control. (c) Western blotting of pRB Ser807/811, total pRB, KLF4 and BRAF in M26c cells transfected with BRAF-V600E for 48 h and treated with 1 μM CI-1040. It shows that BRAF-V600E increases pRB phosphorylation at Ser807/811, which is partially reduced by inhibition of MEK1/2 with CI-1040. HSP90 was used as loading control. (d) Western blotting analysis showing reduced E2F1 and KLF4 expression levels in A375 transduced with LV-shE2F1 compared with control cells (LV-c). Actin was used as loading control. (e) Western blotting analysis of KLF4, E2F1 and pERK1/2 in A375 cells treated with CI-1040 1 μM after transient transfection with E2F1. Actin was used as loading control. (f) KLF4 mRNA levels measured by qPCR in A375 cells treated as indicated. The y axis represents expression ratio of gene/(GAPDH and β-ACTIN average). Data represent mean±s.e.m. of three independent experiments. (g) Chromatin immunoprecipitation (ChIP) assay showing that endogenous E2F1 binds to KLF4 promoter in M26c cells. The y axis represents the relative promoter enrichment, normalized on input material. ACTIN promoter was used as negative control and set to 1. (h) Quantification of dual reporter luciferase assay in M26c melanoma cells showing that E2F1 transactivates KLF4 promoter. Relative luciferase activities were firefly/Renilla ratios, with the level induced by control equated to 1. Data represent mean±s.e.m. of three independent experiments. *P⩽0.05 and **P<0.01.
Converging lines of evidence in different contexts suggest that the RAS/MEK/ERK cascade might activate Cyclin D1/CDK4, which phosphorylate and inactivate the tumor suppressor pRB, releaving inhibition of E2F1 and leading to cell cycle progression.45, 46, 47, 48, 49 To address whether MEK/ERK affects E2F1 also through inactivation of pRB, we tested whether ERK induced phosphorylation of pRB at Ser807/Ser811, the preferential sites of CDK4 phosphorylation.50 Western blot analysis showed that overexpression of BRAF-V600E in M26c cells increased phosphorylation of pRB at Ser807/811 (Figure 3c), whereas CI-1040 treatment decreased it (Figure 3c; Supplementary Figure 4a). These results indicate that MEK/ERK signaling might regulate E2F1, at least in part, through phosphorylation and inactivation of pRB.
To assess whether E2F1 regulates KLF4 expression in melanoma cells, we silenced E2F1 by using a specific short interference RNA (LV-shE2F1).44 Ablation of E2F1 led to a reduction in KLF4 protein levels in A375 cells (Figure 3d). The involvement of E2F1 in the regulation of KLF4 was further confirmed by transiently overexpression of the former in melanoma cells treated with CI-1040. Ectopic expression of E2F1 in melanoma cells strongly enhanced KLF4 expression both at the mRNA and protein levels, whereas the MEK1/2 inhibitor CI-1040 decreased KLF4 level compared with control. Interestingly, overexpression of E2F1 in presence of CI-1040 was able to revert the effect of the inhibitor maintaining high levels of KLF4 (Figures 3e and f; Supplementary Figure S4b).
A web-based bioinformatic analysis identified a putative E2F1 binding site within the human KLF4 promoter (TTTCTCGC), which is identical to the canonical E2F1 consensus sequence,51 except for the presence of a Thymine instead of Cytosine in position 5 (Figure 3g). Therefore, we hypothesized KLF4 as a direct transcriptional target of E2F1. Nuclear chromatin immunoprecipitation of endogenous E2F1 in M26c melanoma cells showed binding to KLF4 promoter at 1204 bp from the transcription start site (Figure 3g). To demonstrate that the binding of E2F1 to KLF4 promoter is functionally active, we cloned 2.7 Kb of the KLF4 promoter upstream to a luciferase gene and we co-transfected this reporter together with an expression plasmid of E2F1. We found that E2F1 strongly induced KLF4 promoter activity (Figure 3h), indicating the presence of a functional E2F1-binding site in KLF4 promoter. Altogether, these data suggest that in melanoma cells KLF4 is positively modulated by the RAS/RAF/MEK/ERK signaling through E2F1.
KLF4 increases melanoma cell growth and reduces apoptosis
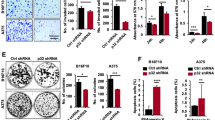
To assess the role of KLF4 in melanoma, we ectopically expressed KLF4 by lentivirus-mediated delivery in two patient-derived melanoma cells expressing low/moderate KLF4 levels (M26c, M33x). Stable overexpression of FL KLF4, confirmed at protein level (Figure 4a), induced a time-dependent increase in melanoma cell growth (Figure 4b) and enhanced colony formation in soft agar (Figure 4c). KLF4 overexpression drastically reduced the fraction of early and late apoptotic cells, as revealed by Annexin V/7AAD staining (Figure 4d). The anti-apoptotic function of KLF4 in melanoma cells was confirmed by the decrease of total Caspase-3 and increase of the anti-apoptotic factor BCL-2 upon stable transfection of FL KLF4 (Figure 4e). Consistently, qPCR analysis showed increased mRNA levels of BCL-2 and BCL-XL and decreased levels of BAX compared with cells transfected with empty vector (Figure 4f). These results indicate that KLF4 enhances melanoma cell growth by decreasing apoptosis.
Ectopic expression of KLF4 promotes cell growth and affects apoptosis in melanoma cells. (a) Western blotting analysis of KLF4 in M26c and M33x cells stably transfected with LV-KLF4. Actin was used as loading control. (b) Cell viability assay in M26c and M33x stably transfected with LV-KLF4 and control LV-c. Data shown are mean±s.e.m. of at least three biological replicates, each performed in triplicate. (c) Histogram of the quantification and representative images of the soft agar assay in M33x cells stably transfected with LV-c and LV-KLF4. The y axis represents colony number. Data shown are mean±s.e.m. of at least three biological replicates, each performed in triplicate. (d) M26c and M33x melanoma cells stably transfected as indicated, were subjected to cytometric analysis of apoptotic cells after staining with Annexin V/7-AAD (Annexin V+/7-AAD−: early apoptosis; Annexin V+/7-AAD+: late apoptosis). The data represent mean±s.e.m. of three independent experiments. (e) Western blotting analysis of KLF4, BCL2 and Caspase-3 in M26c and M33x stably transfected as described above. Actin was used as loading control. (f) qPCR analysis of BCL2, BCL-XL and BAX in M26c cells stably transfected with KLF4. The y axis represents expression ratio of gene/(GAPDH and β-ACTIN average), with the control equated to 1. Data shown are mean±s.e.m. of at least three biological replicates, each performed in triplicate. *P⩽0.05, **P<0.01.
FL KLF4 and the splice variant α have shown to exert opposite effects in pancreatic cancer cells.16, 29 To test whether this behavior occurred in melanoma, KLF4α was stably transfected in M26c cells (Supplementary Figure S5a). Overexpression of KLF4α promoted melanoma cell growth and decreased the fraction of early and late apoptotic cells (Supplementary Figures S5b and c), at the same extent as KLF4 FL (Figure 4b). These results indicate that both KLF4 FL and the isoform α exert a pro-tumorigenic role in melanoma cells. The lack of nuclear localization signal and DNA-binding domain in KLF4α infers that this isoform by itself is not able to enter the nucleus and promote cell growth. Consistently with this, co-immunoprecipitation experiments showed that when Myc-tagged KLF4α was overexpressed in melanoma cells, endogenous KLF4 immunoprecipitated with the exogenous isoform α (Supplementary Figure S5d), confirming a physical interaction between KLF4 FL and KLF4α, as previously reported.16 Interestingly, we found that overexpression of KLF4α in absence of KLF4 FL was unable to increase melanoma cell proliferation and had the same effect as KLF4 silencing, suggesting that the isoform KLF4α requires KLF4 FL to elicit its pro-tumorigenic function (Supplementary Figure S5e).
KLF4 silencing reduces melanoma cell proliferation and induces apoptosis
To further elucidate the role of KLF4, we tested the effect of KLF4 silencing in melanoma cells with high (A375) and moderate (M26c) KLF4 levels using two specific short interference RNAs (LV-shKLF4-1, LV-shKLF4-2), that resulted in 80–90% reduction of KLF4 protein and mRNA levels (Figure 5a; Supplementary Figure S6a). KLF4 silencing did not affect the expression of KLF5 mRNA (Supplementary Figure S6b), suggesting that KLF5 does not compensate for ablation of KLF4. Silencing of KLF4 with both short interference RNAs decreased melanoma cell growth and colony formation compared with the control vector (LV-c) (Figures 5b and c). The role of KLF4 in controlling melanoma cell proliferation was also confirmed by analysis of the proliferation index, determined by carboxyfluorescein succinimidyl ester staining. The assay showed that A375 and M26c KLF4-depleted cells grew slower than control cells (Figure 5d). Cell cycle analysis did not reveal any significant changes (Supplementary Figure S6c). KLF4 silencing increased the fraction of early and late apoptotic cells in both cell types, as revealed by Annexin V/7AAD staining (Figure 5e). Similar results were observed after propidium iodide staining (Supplementary Figure S6d). The effect of KLF4 silencing on apoptosis was associated with an upregulation of p53, BAX and cleaved Caspase-3, and induction of poly ADP-ribose polymerase cleavage in cells transduced with LV-shKLF4 (Figure 5f). qPCR analysis in melanoma cells revealed that depletion of KLF4 decreased mRNA levels of the anti-apoptotic markers BCL-2 and BCL-XL, and increased the pro-apoptotic factor NOXA (Figure 5g). Altogether, these results suggest that silencing of KLF4 reduces melanoma cell proliferation and induces apoptosis. As a further control, we modulated KLF4 expression in two cancer cell lines in which KLF4 is known to act as a tumor suppressor.52, 53 As expected, KLF4 overexpression decreased proliferation in the chronic myeloid leukemia cell line K562, whereas its silencing increased cell number in K562 cells and in the colorectal cancer cell line HCT116 (Supplementary Figure S7).
Knockdown of KLF4 inhibits cell growth and increases apoptosis. (a) Western blotting analysis of KLF4 in A375 and M26c cells transduced with LV-c, LV-shKLF4-1 or LV-shKLF4-2. Actin was used as loading control. (b) Growth curve in A375 and M26c cells transduced as indicated. Data shown are mean±s.e.m. of at least three biological replicates, each performed in triplicate. (c) Histogram of the quantification and representative images of the colony assay in A375 cells transduced with LV-c, LV-shKLF4-1 or LV-shKLF4-2 lentivectors. The y axis represents percentage of colony number, with control set to 100%. Data shown are mean±s.e.m. of at least three biological replicates, each performed in triplicate. (d) Proliferation index measured by carboxyfluorescein succinimidyl ester staining in A375 and Me26c melanoma cells stably transduced with LV-c, LV-shKLF4-1 or LV-shKLF4-2. Data shown are mean±s.e.m. of at least three biological replicates, each performed in triplicate. (e) A375 and M26c melanoma cells transduced as indicated, were subjected to cytometric analysis of apoptotic cells after staining with Annexin V/7-AAD (Annexin V+/7-AAD−: early apoptosis; Annexin V+/7-AAD+: late apoptosis). The data represent mean±s.e.m. of three independent experiments. (f) Western blot analysis of poly ADP-ribose polymerase (PARP), BAX, p53 and Cleaved Caspase-3 in A375 and M26c cells transduced as indicated. Actin served as loading control. (g) Quantitative PCR analysis of BCL2, BCL-XL and NOXA expression in A375 and M26c cells transduced as indicated. The y axis represents expression ratio of gene/(GAPDH and β-ACTIN average), with control equated to 1. Data represent mean±s.e.m. of three independent experiments. (h) Quantification of p53-dependent luciferase reporter assay (p21-Luc) to test the efficacy of KLF4 silencing alone or in combination with shp53. The data represent mean±s.e.m. of three independent experiments. *P⩽0.05 and **P<0.01.
The molecular mechanism underlying the increased cell death induced by KLF4 depletion might be due to activation of p53 activity.15 To address this hypothesis, we performed a luciferase assay to detect p21 transcriptional activity after silencing of KLF4 and p53, alone or in combination. Our data indicate that silencing of KLF4 increased the transcriptional activity of p21, a direct target of p53. This effect was abolished when KLF4 was silenced in combination with silencing of p53, suggesting a p53-dependent mechanism (Figure 5h). To test whether the effect of KLF4 on cell death is due to p53, we silenced KLF4 and p53 in A375 and Mewo cells, which harbor, respectively, wt and mutant p53. In A375 cells silencing of p53 completely abolished the effect on early apoptosis induced by KLF4 depletion. In Mewo cells silencing of p53 only slight decreased the effect of KLF4 knock-down on early apoptosis (Supplementary Figure S8). Taken together, these data indicate that the apoptotic effect induced by depletion of KLF4 is not exclusively dependent on p53.
KLF4 is required for melanoma xenograft growth
To investigate whether KLF4 might regulate melanoma xenograft growth in vivo, A375 cells stably transduced with LV-c or LV-shKLF4 were subcutaneously injected into athymic nude mice, and tumor growth was monitored. KLF4 silencing reduced of about 50% the size of melanoma xenografts compared with LV-c (Figures 6a and b). Western blot analysis of dissected tumors confirmed the reduction of KLF4 levels upon KLF4 silencing (Figure 6c). The degree of reduction of melanoma xenograft growth was consistent with the decrease of melanoma cell growth observed in vitro. Altogether, these results indicate that KLF4 is required for melanoma cell growth in vitro and in vivo, further confirming the oncogenic function of KLF4 in melanoma.
KLF4 is required for melanoma cell growth in vivo. (a) A375 cells transduced with LV-c and LV-shKLF4 lentiviruses were injected subcutaneously in athymic-nude mice. Quantification of tumor volume (n=6 per group), showing that KFL4 silencing prevented the increase of tumor growth. (b) Representative images of A375 xenografts, as indicated. (c) Western blotting analysis of KLF4 in tumors derived from A375 xenografts. Actin served as loading control. (d) Growth assay in M26c cells transiently transfected with LV-c, LV-shKLF4, pBRAF-V600E and LV-shKLF4+pBRAFV600E. (e) Western blotting analysis of KLF4, BRAF, pERK1/2 in M26c cells transfected as indicated. Actin was used as loading control. *P⩽0.05 and **P<0.01.
Finally, we tested the role of KLF4 in the growth promoting effect mediated by activation of the RAS/RAF/MEK/ERK signaling. Interestingly, we found that KLF4 silencing drastically affected BRAF-mediated increased proliferation in M26c cells, as indicated by 50% reduction in proliferation rate of BRAF-V600E/shKLF4 cells compared with transfected with BRAF-V600E alone (Figures 6d and e). These results suggest that KLF4 is required to maintain BRAF-mediated melanoma cell growth.
Discussion
The majority of cutaneous melanomas present aberrant activation of the RAS/RAF/MEK/ERK signaling pathway, which is critical for melanoma growth and maintenance.2, 3, 4 In this study, we uncover the pro-oncogenic role of KLF4 in human melanoma and we provide evidence that KLF4 is positively modulated by the RAS/RAF/MEK/ERK signaling through the transcription factor E2F1. Collectively, these findings identify KLF4 as an important factor that regulates melanoma cell growth downstream of ERK, suggesting that KLF4 might be a potential therapeutic target in melanoma.
KLF4 can have a dual role in tumorigenesis, acting either as a tissue-specific tumor suppressor or oncogene, likely due to its ability to induce cell cycle arrest and/or block apoptosis.14 Several lines of evidence support the tumor-suppressive function of KLF4 in several types of cancer, including gastric, colon, lung and renal cancers, and leukemia.19, 20, 21, 22, 23 A phase 1 study with APTO-253, an inducer of KLF4, demonstrated modest clinical activity in patients with advanced or metastatic solid tumors.24 However, KLF4 was also reported to have a potent oncogenic function in osteosarcoma25 and in breast cancer, where it is required for the maintenance of cancer stem cells and for cell migration and invasion.26 Forced expression of KLF4 in basal keratinocytes can initiate squamous epithelial dysplasia.30 Furthermore, recent studies have revealed a more complex scenario, in which KLF4 can act with opposite stage-dependent functions in PDA and in esophageal squamous carcinoma.28, 54
Recent microarray data revealed that KLF4 expression is elevated in melanoma cell lines compared with melanocytes;55 however, it remains unknown whether and how KLF4 functions in this tumor. Our findings provide several lines of evidence supporting the pro-oncogenic role of KLF4 in melanoma. First, KLF4 ablation drastically reduces melanoma cell proliferation in vitro and melanoma xenograft growth in vivo. Second, KLF4 is required to maintain oncogenic BRAF-mediated melanoma cell growth in vitro. Third, enhanced KLF4 expression increases melanoma cell growth and colony formation. Our data indicate that KLF4 promotes tumor growth by decreasing apoptosis, as revealed by downregulation of p53, p21 and BAX, and upregulation of the anti-apoptotic factors BCL-2 and BCL-XL. These findings are consistent with a previous report showing that KLF4 depletion restores p53 and results in p53-dependent apoptosis.15 Although p53 is a well known target of KLF4, that mediates the apoptosis induced by KLF4 silencing in melanoma, our data suggest that KLF4 can also induce p53-independent cell death. The pathways involved in such effect remain unknown at the moment and further studies in p53 mutated melanoma cells are warranted to elucidate this effect.
Another interesting finding of this study is the identification of a biologically active KLF4α isoform, which is expressed at various levels in the majority of melanomas. Our data indicate that KLF4α promotes cell proliferation and decreases apoptosis (Supplementary Figures S5b and 5c), suggesting that both KLF4 and its splice variant KLF4α exert a pro-tumorigenic role in melanoma cells. These results are at variance with what reported in pancreatic cancer cells, where KLF4 and KLF4α have opposite functions; FL KLF4 inhibits cell growth and is associated with blockage of cell cycle progression and upregulation of p27 and p21,29 whereas KLF4α promotes cell cycle progression and reduces the expression of p27 and p21.16 KLF4α protein lacks the nuclear localization signal and the DNA-binding domain, is mostly localized in the cytoplasm, and partially retains the N-terminal transcription activation domain of the FL, whereas KLF4 is mainly localized and functions in the nucleus. Therefore, since KLF4 and the isoform α physically interact, it is reasonable to speculate that the pro-oncogenic function of KLF4α is due to this interaction and that KLF4α requires KLF4 FL to exert its pro-tumorigenic function in melanoma cells. This is supported by our data showing that KLF4α overexpression in absence of KLF4 is unable to promote melanoma cell proliferation and has the same effect as KLF4 silencing.
The most common oncogenic events in melanomas are mutually exclusive activating mutations in BRAF and NRAS, which occur in 75% of cases.2, 3, 4 Activating mutations in BRAF/NRAS and other genetic events cause constitutive activation of the downstream ERK1/2, resulting in pro-proliferative and anti-apoptotic effects that promote tumor cell growth. Here, we find that the RAS/RAF/MEK/ERK signaling positively modulates the expression of KLF4 and that KLF4 is required to maintain the proliferative effect of oncogenic BRAF in melanoma cells. Interestingly, modulation of the RAS/RAF/MEK/ERK signaling does not affect the expression of KLF5, a closely related member of the KLF family, which we found to be correlated with KLF4 in melanoma cells. These findings suggest that in melanoma KLF4 is the main mediator of the RAS/RAF/MEK/ERK pathway, despite KLF5 has been shown to mediate KRAS-induced intestinal tumorigenesis.56
Our data suggest that the induction of KLF4 expression is mediated by the transcription factor E2F1, which binds the KLF4 promoter (TTTCTCGC) in a binding site that differs from the canonical E2F1 sequence for the presence of a Thymine instead of Cytosine in position 5th. The biological relevance of the regulation of KLF4 by E2F1 is supported by a positive correlation between the expression of KLF4 and E2F1 in melanoma tissues. Our data suggest that ERK1/2 modulates both activity and expression of E2F1, possibly through two non-exclusive mechanisms (Figure 7). First, mutated BRAF increases CDK4/CyclinD1-dependent pRB phosphorylation, and this effect is mediated by ERK1/2 (Figure 3c). It is, therefore, reasonable to assume that phosphorylation of pRB leads to its inactivation, with consequent destabilization of the pRB/E2F1 interaction, and release of E2F1, which is free to induce transcription of KLF4. Second, our data suggest that ERK1/2 modulates the expression of E2F1, as indicated by a strong reduction of E2F1 protein level upon MEK1/2 inhibition (Figure 3). At present it remains unclear whether ERK1/2 modulates the expression of E2F1 protein directly, since ERK1/2 has also a non-catalytic activity,57 or indirectly through a third protein, which, in turn, modulates E2F1.
Model of the ERK1/2-E2F1-KLF4 axis. E2F1 acts downstream of ERK1/2 to regulate the expression of KLF4 by direct binding to KLF4 promoter. ERK1/2 can regulate E2F1 via CycD1/CDK4, which in turn promotes phosphorylation of pRB and consequent release of E2F1. Alternatively ERK1/2 can directly or indirectly regulate E2F1 expression. CI-1040 is a MEK1/2 inhibitor and SCH772984 is a ERK1/2 inhibitor.
In summary, this study provides mechanistic evidence for a pro-tumorigenic role of KLF4 in melanoma and uncovers a novel ERK1/2-E2F1-KLF4 axis that regulates melanoma cell growth, suggesting that aberrant KLF4 may be a potential therapeutic target in a subset of melanomas. Further studies are needed to explore novel approaches to effectively and safely target KLF4 in patients. Our data indicate that KLF5 does not compensate for loss of KLF4. However, it will be important to assess the compensatory effects of other members of the KLF family, which might impair the efficacy of potential therapies targeting KLF4.
Material and methods
Cell lines and melanoma samples
A375 (CRL-1619) and Mewo (HTB-65) melanoma cells were purchased at American Type Culture Collection (Manassas, VA, USA) and normal human epidermal melanocytes at PromoCell (Heidelberg, Germany). SK-Mel-2, SK-Mel-5 and SK-Mel-28 melanoma cells were kindly provided by Dr Laura Poliseno (CRL-ITT, Pisa, Italy). Cell lines were authenticated by short tandem repeat profiling. Human melanoma samples (Supplementary Table S1) were obtained after approved protocols by the local Ethics Committee, and processed as previously described.7, 8 Patient-derived SSM2c, M26c, M33x melanoma cells were already described8 and M51 cells were derived from a subcutaneous melanoma metastasis. Lentiviruses were produced in HEK-293T cells as already described.58 Cells were periodically tested for Mycoplasma contamination by 4’,6-diamidino-2-phenylindole inspection and PCR. CI-1040 and SCH772984 (Sigma, St Louis, MO, USA) were used for 16 h, respectively, at 1 μM and 0.5 μM, and transduced cells were selected with puromycin (2 μg/ml) (Sigma).
Cell growth, colony formation assay and soft agar
For growth curves, 3000 cells/well were plated in 12-well plates and counted on days 3, 5 and 7. For colony formation assay, 500 cells/well were plated in six-well plates. After 10 days, cells were fixed with methanol, stained with Crystal Violet and colonies counted. For soft agar assay, cells were suspended in medium containing 0.5% agar/well and overlaid on 2% agar in six-well plates (5000 cells/well). After two weeks cells were fixed as described above and counted.
Plasmid construction and RNA interference
Full-length KLF4 and the isoform KLF4alpha were PCR amplified with Platinum Pfx DNA polymerase (Life Technologies, Grand Island, NY, USA) and cloned, respectively, into LV-pBABE and pCS2+MT (Addgene, Cambridge, MA, USA) vectors. PCR primers used to cover both 5'- and 3'-untranslated regions of KLF4 were as follow: forward primer 5′-AAAAAAGAATTCATGAGGCAGCCACCTG-3′ and reverse primer 5′-AAAAAAGTCGACTTAAAAATGCCTCTTCATGTG-3'. KLF4 promoter (-2731) was PCR amplified with Platinum Pfx polymerase (Life Technologies) and cloned into pGL3Basic vector (Promega, Madison, WI, USA) using SacI-HindIII sites, to generate KLF4 prom-luc reporter. Primers used were KLF4 prom FW: 5′-AAAAAGAGCTCTCCTTTTTTGGTTCCTCCGTT-3′, KLF4 prom RV: 5′-AAAAAAAGCTTAAAGTTCTTAGAAAAGTTGTAA-3′. Lentiviruses were produced in HEK-293T cells. Lentiviral vectors used were: pLKO.1-puro (LV-c), pLKO.1-puro-shKLF4-1 (LV-shKLF4-1) (targeting sequence 5′-GCCTTACACATGAAGAGGCAT-3′), pLKO.1-puro-shKLF4-2 (LV-shKLF4-2) (targeting sequence 5′-TTGTGGATATCAGGGTATAAA-3′). pCDNA3.1-BRAF wt and pCDNA3.1-BRAF-V600E constructs were kindly provided by L. Poliseno. Transfections were performed in OptiMEM (Life Technologies, Carlsbad, CA, USA) using X-tremeGENE transfection reagent (Roche Diagnostic, Milan, Italy).
Immunofluorescence
Melanoma cells were fixed with 4% paraformaldehyde and incubated with rabbit anti-KLF4 (sc-20691) (Santa Cruz Biotechnologies, Santa Cruz, CA, USA) and mouse anti-Myc (9E10, Santa Cruz Biotechnology) antibodies. Secondary antibodies were anti-rabbit fluorescein isothiocyanate-conjugated and anti-mouse Rhodamine-conjugated (Life Technologies). Cells were counterstained with 4’,6-diamidino-2-phenylindole. Immunofluorescence was visualized with a Zeiss Observer.z1.
Western Blotting, co-immunoprecipitation and cell fractionation
Cells were lysed in 20 mM Hepes buffer, 10 mM KCl, 1 mM EDTA, 0.2% NP-40, 10% Glycerol44, 59 and co-immunoprecipitation studies were performed as previously described.44 Details can be found in Supplementary Information. The following antibodies were used: mouse monoclonal anti-β-Actin (AC-15, Sigma-Aldrich, St Louis, MO, USA), mouse anti-HSP90 (Heat Shock Protein 90), rabbit polyclonal anti-KLF4 (sc-20691), rabbit polyclonal anti-p21 (C-19; sc-397), mouse monoclonal anti-p53 (DO-1; sc-126), mouse anti-Myc (9E10), mouse monoclonal anti-Caspase-3 (E-8; sc-7272), goat anti-Fibrillarin (D-14; sc-11336), goat anti-GAPDH (V-18; sc-20357), and mouse anti-BRAF wt (sc-5284) (Santa Cruz Biotechnology), rabbit polyclonal anti-BAX, rabbit polyclonal anti-BCL2, rabbit polyclonal anti-E2F1, rabbit polyclonal anti-poly ADP-ribose polymerase, rabbit polyclonal anti-Phospho-p44/42 MAPK (ERK1/2) (Thr202/Tyr204), rabbit polyclonal anti-Cleaved Caspase 3 (Asp175) (Cell Signaling Technologies, Danvers, MA, USA). Chemiluminescent detection was used.
RNA isolation and qPCR
Total RNA was isolated as already described.44 Details can be found in Supplementary Information. Primer sequences are listed in Supplementary Table 2.
Luciferase reporter assay
A p53-responsive (p21-Luc) luciferase reporter (Addgene)60 was used in combination with Renilla luciferase pRL-TK reporter vector (Promega) (ratio 10:1) to normalize luciferase activity as already described.58 pGL3Basic vector (Promega) was used to equal DNA amounts. Luminescence was quantified using the Dual-Glo Luciferase Assay System (Promega) and the GloMax 20/20 Luminometer (Promega).
Nuclear chromatin immunoprecipitation experiments
M26c cells were fixed with 1% formaldehyde and lysed. DNA was sonicated and diluted with chromatin immunoprecipitation dilution buffer, and input material (5%) was collected. Chromatin was incubated overnight at 4 °C with Dynabeads Protein G (Life Technologies) pre-conjugated with anti-E2F1 (Cell Signaling) or a non-specific IgG control. DNA was purified, and qPCRs was carried out at 60 °C using Power SYBR Green PCR Master Mix (Life Technologies). Chromatin immunoprecipitation primers are reported in Supplementary Table 2.
Flow cytometry analysis
For proliferation index, cells were labeled with 5 μM of CellTrace Violet (Life Technologies), seeded and allowed to proliferate for 72, 96 and 120 h and analyzed using flow cytometry. CellTrace Violet data were normalized to controls arrested at the parent generation with 1 μg/ml mitomycin C (t=0 h) and proliferation index was calculated using ModFit LT software (Verity Software House, Topsham, ME, USA). For evaluation of cell death, cells were serum-deprived for 48 h and analyzed with an Annexin V/7-AAD apoptosis kit (BD Biosciences, San Diego, CA, USA). Cytometric analysis was performed with FACS-Canto II (Becton Dickinson, Franklin Lakes, NJ, USA).
Xenografts
A375 cells were transduced with LV-c or LV-shKLF4, resuspended in Matrigel (Becton Dickinson)/Dulbecco's Modified Eagle's medium (1/1) and subcutaneously injected into lateral flanks of adult female athymic-nude mice (Foxn1 nu/nu) (Harlan Laboratories, Udine, Italy) (10 000 cells/injection). Mice were randomly chosen for the two experimental groups and housed in specific pathogen free conditions. Subcutaneous tumor size was blindly measured twice a week with a caliper. Tumor volumes were calculated using the formula: V=W2x L x 0.5, where W is tumor width and L is tumor length. No animals were excluded from the analysis. No statistical methods were used for sample size estimation. The experiment was approved by the Italian Ministry of Health and was in accordance with the Italian guidelines and regulations.
Statistical analysis
Data are presented as mean±s.e.m. for at least three independent experiments, unless otherwise stated. No statistical methods were used for sample size selection. The estimate of variation within each group was similar. Experiments were either analyzed by using a two-tailed Student’s t-test or one-way analysis of variance followed by Bonferroni’s post-hoc test for multiple comparisons. *P⩽0.05; **P⩽0.01.
References
Balch CM, Gershenwald JE, Soong SJ, Thompson JF, Atkins MB, Byrd DR et al2009. Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol 2009; 27: 6199–6206.
Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S et al. Mutations of the BRAF gene in human cancer. Nature 2002; 417: 949–954.
Brose MS, Volpe P, Feldman M, Kumar M, Rishi I, Gerrero R et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res 2002; 62: 6997–7000.
Tsao H, Chin L, Garraway LA, Fisher DE . Melanoma: from mutations to medicine. Genes Dev 2012; 26: 1131–1155.
Lo JA, Fisher DE . The melanoma revolution: from UV carcinogenesis to a new era in therapeutics. Science 2014; 346: 945–949.
Stecca B, Mas C, Clement V, Zbinden M, Correa R, Piguet V et al. Melanomas require HEDGEHOG-GLI signaling regulated by interactions between GLI1 and the RAS-MEK/AKT pathways. Proc Natl Acad Sci USA 2007; 104: 5895–5900.
Santini R, Vinci MC, Pandolfi S, Penachioni JY, Montagnani V, Olivito B et al. Hedgehog-GLI signaling drives self-renewal and tumorigenicity of human melanoma-initiating cells. Stem Cells 2012; 30: 1808–1818.
Santini R, Pietrobono S, Pandolfi S, Montagnani V, D'Amico M, Penachioni JY et al. SOX2 regulates self-renewal and tumorigenicity of human melanoma-initiating cells. Oncogene 2014; 33: 4697–4708.
Segre JA, Bauer C, Fuchs E . Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet 1999; 22: 356–360.
Shields JM, Christy RJ, Yang VW . Identification and characterization of a gene encoding a gut-enriched Krüppel-like factor expressed during growth arrest. J Biol Chem 1996; 271: 20009–20017.
Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007; 131: 861–872.
Utikal J, Maherali N, Kulalert W, Hochedlinger K . Sox2 is dispensable for the reprogramming of melanocytes and melanoma cells into induced pluripotent stem cells. J Cell Sci 2009; 122: 3502–3510.
El-Karim EA, Hagos EG, Ghaleb AM, Yu B, Yang VW . Krüppel-like factor 4 regulates genetic stability in mouse embryonic fibroblasts. Mol Cancer 2013; 12: 89.
Tetreault MP, Yang Y, Katz JP . Krüppel-like factors in cancer. Nat Rev Cancer 2013; 13: 701–713.
Rowland BD, Bernards R, Peeper DS . The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat Cell Biol 2005; 7: 1074–1082.
Wei D, Wang L, Kanai M, Jia Z, Le X, Li Q et al. KLF4α up-regulation promotes cell cycle progression and reduces survival time of patients with pancreatic cancer. Gastroenterology 2010; 139: 2135–2145.
Gamper AM, Qiao X, Kim J, Zhang L, DeSimone MC, Rathmell WK et al. Regulation of KLF4 turnover reveals an unexpected tissue-specific role of pVHL in tumorigenesis. Mol Cell 2012; 45: 233–243.
Kim MO, Kim SH, Cho YY, Nadas J, Jeong CH, Yao K et al. ERK1 and ERK2 regulate embryonic stem cell self-renewal through phosphorylation of Klf4. Nat Struct Mol Biol 2012; 19: 283–290.
Katz JP, Perreault N, Goldstein BG, Actman L, McNally SR, Silberg DG et al. Loss of Klf4 in mice causes altered proliferation and differentiation and precancerous changes in the adult stomach. Gastroenterology 2005; 128: 935–945.
Wei D, Gong W, Kanai M, Schlunk C, Wang L, Yao JC et al. Drastic down-regulation of Krüppel-like factor 4 expression is critical in human gastric cancer development and progression. Cancer Res 2005; 65: 2746–2754.
Yu T, Chen X, Zhang W, Liu J, Avdiushko R, Napier DL et al. KLF4 regulates adult lung tumor-initiating cells and represses K-Ras-mediated lung cancer. Cell Death Differ 2016; 23: 207–215.
Li H, Wang J, Xiao W, Xia D, Lang B, Yu G et al. Epigenetic alterations of Krüppel-like factor 4 and its tumor suppressor function in renal cell carcinoma. Carcinogenesis 2013; 34: 2262–2270.
Kharas MG, Yusuf I, Scarfone VM, Yang VW, Segre JA, Huettner CS et al. KLF4 suppresses transformation of pre-B cells by ABL oncogenes. Blood 2007; 109: 747–755.
Cercek A, Wheler J, Murray PE, Zhou S, Saltz L . Phase 1 study of APTO-253 HCl, an inducer of KLF4, in patients with advanced or metastatic solid tumors. Invest New Drugs 2015; 33: 1086–1092.
Zhang L, Zhang L, Xia X, He S, He H, Zhao W . Krüppel-like factor 4 promotes human osteosarcoma growth and metastasis via regulating CRYAB expression. Oncotarget 2016; 7: 30990–31000.
Yu F, Li J, Chen H, Fu J, Ray S, Huang S et al. Kruppel-like factor 4 (KLF4) is required for maintenance of breast cancer stem cells and for cell migration and invasion. Oncogene 2011; 30: 2161–2172.
Yori JL, Seachrist DD, Johnson E, Lozada KL, Abdul-Karim FW, Chodosh LA et al. Krüppel-like factor 4 inhibits tumorigenic progression and metastasis in a mouse model of breast cancer. Neoplasia 2011; 13: 601–610.
Wei D, Wang L, Yan Y, Jia Z, Gagea M, Li Z et al. KLF4 is essential for induction of cellular identity change and acinar-to-ductal reprogramming during early pancreatic carcinogenesis. Cancer Cell 2016; 29: 324–338.
Wei D, Kanai M, Jia Z, Le X, Xie K . Kruppel-like factor 4 induces p27Kip1 expression in and suppresses the growth and metastasis of human pancreatic cancer cells. Cancer Res 2008; 68: 4631–4639.
Foster KW, Liu Z, Nail CD, Li X, Fitzgerald TJ, Bailey SK et al. Induction of KLF4 in basal keratinocytes blocks the proliferation-differentiation switch and initiates squamous epithelial dysplasia. Oncogene 2005; 24: 1491–1500.
Li J, Zheng H, Yu F, Yu T, Liu C, Huang S, Wang TC, Ai W . 2012. Deficiency of the Kruppel-like factor KLF4 correlates with increased cell proliferation and enhanced skin tumorigenesis. Carcinogenesis 33: 1239–1246.
Camacho-Vanegas O, Till J, Miranda-Lorenzo I, Ozturk B, Camacho SC, Martignetti JA . Shaking the family tree: identification of novel and biologically active alternatively spliced isoforms across the KLF family of transcription factors. FASEB J 2013; 27: 432–436.
Farrugia MK, Sharma SB, Lin CC, McLaughlin SL, Vanderbilt DB, Ammer AG et al. Regulation of anti-apoptotic signaling by Kruppel-like factors 4 and 5 mediates lapatinib resistance in breast cancer. Cell Death Dis 2015; 6: e1699.
Evans PM, Liu C . Roles of Krüpel-like factor 4 in normal homeostasis, cancer and stem cells. Acta Biochim Biophys Sin (Shanghai) 2008; 40: 554–564.
Platz A, Egyhazi S, Ringborg U, Hansson J . Human cutaneous melanoma; a review of NRAS and BRAF mutation frequencies in relation to histogenetic subclass and body site. Mol Oncol 2008; 1: 395–405.
Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet 2012; 44: 1006–1014.
Montagnani V, Benelli M, Apollo A, Pescucci C, Licastro D, Urso C et al. Thin and thick primary cutaneous melanomas reveal distinct patterns of somatic copy number alterations. Oncotarget 2016; 7: 30365–30378.
Delaney AM, Printen JA, Chen H, Fauman EB, Dudley DT . Identification of a novel mitogen-activated protein kinase kinase activation domain recognized by the inhibitor PD 184352. Mol Cell Biol 2002; 22: 7593–7602.
Nikolaev SI, Rimoldi D, Iseli C, Valsesia A, Robyr D, Gehrig C et al. Exome sequencing identifies recurrent somatic MAP2K1 and MAP2K2 mutations in melanoma. Nat Genet 2011; 44: 133–139.
Morris EJ, Jha S, Restaino CR, Dayananth P, Zhu H, Cooper A et al. Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer Discov 2013; 3: 742–750.
Filmus J, Robles AI, Shi W, Wong MJ, Colombo LL, Conti CJ . Induction of cyclin D1 overexpression by activated ras. Oncogene 1994; 9: 3627–3633.
Peeper DS, Upton TM, Ladha MH, Neuman E, Zalvide J, Bernards R et al. Ras signalling linked to the cell-cycle machinery by the retinoblastoma protein. Nature 1997; 386: 177–181.
Pützer BM, Steder M, Alla V . Predicting and preventing melanoma invasiveness: advances in clarifying E2F1 function. Expert Rev Anticancer Ther 2010; 10: 1707–1720.
Pandolfi S, Montagnani V, Lapucci A, Stecca B . HEDGEHOG/GLI-E2F1 axis modulates iASPP expression and function and regulates melanoma cell growth. Cell Death Differ 2015; 22: 2006–2019.
Mittnacht S, Paterson H, Olson MF, Marshall CJ . Ras signalling is required for inactivation of the tumour suppressor pRb cell-cycle control protein. Curr Biol 1997; 7: 219–221.
Mittnacht S . Control of pRB phosphorylation. Curr Opin Genet Dev 1998; 8: 21–27.
Garnovskaya MN, Mukhin YV, Vlasova TM, Grewal JS, Ullian ME, Tholanikunnel BG et al. Mitogen-induced rapid phosphorylation of serine 795 of the retinoblastoma gene product in vascular smooth muscle cells involves ERK activation. J Biol Chem 2004; 279: 24899–24905.
Korotayev K, Chaussepied M, Ginsberg D . ERK activation is regulated by E2F1 and is essential for E2F1-induced S phase entry. Cell Signal 2008; 20: 1221–1226.
Rubin SM . Deciphering the retinoblastoma protein phosphorylation code. Trends Biochem Sci 2013; 38: 12–19.
Driscoll B, T'Ang A, Hu YH, Yan CL, Fu Y, Luo Y et al. Discovery of a regulatory motif that controls the exposure of specific upstream cyclin-dependent kinase sites that determine both conformation and growth suppressing activity of pRb. J Biol Chem 1999; 274: 9463–9471.
Rabinovich A, Jin VX, Rabinovich R, Xu X, Farnham PJ . E2F in vivo binding specificity: comparison of consensus versus nonconsensus binding sites. Genome Res 2008; 18: 1763–1777.
Huang Y, Chen J, Lu C, Han J, Wang G, Song C et al. HDAC1 and Klf4 interplay critically regulates human myeloid leukemia cell proliferation. Cell Death Dis 2014; 5: e1491.
Yoon HS, Chen X, Yang VW . Kruppel-like factor 4 mediates p53-dependent G1/S cell cycle arrest in response to DNA damage. J Biol Chem 2003; 278: 2101–2105.
Yang Y, Katz JP . KLF4 is downregulated but not mutated during human esophageal squamous cell carcinogenesis and has tumor stage-specific functions. Cancer Biol Ther 2016; 17: 422–429.
Muthusamy V, Premi S, Soper C, Platt J, Bosenberg M . The hematopoietic stem cell regulatory gene latexin has tumor-suppressive properties in malignant melanoma. J Invest Dermatol 2013; 133: 1827–1833.
Nandan MO, McConnell BB, Ghaleb AM, Bialkowska AB, Sheng H, Shao J et al. Krüppel-like factor 5 mediates cellular transformation during oncogenic KRAS-induced intestinal tumorigenesis. Gastroenterology 2008; 134: 120–130.
Hong SK, Yoon S, Moelling C, Arthan D, Park JI . Noncatalytic function of ERK1/2 can promote Raf/MEK/ERK-mediated growth arrest signaling. J Biol Chem 2009; 284: 33006–33018.
Stecca B, Ruiz i Altaba A . A GLI1-p53 inhibitory loop controls neural stem cell and tumour cell numbers. EMBO J 2009; 28: 663–676.
Pandolfi S, Montagnani V, Penachioni JY, Vinci MC, Olivito B, Borgognoni L et al. WIP1 phosphatase modulates the Hedgehog signaling by enhancing GLI1 function. Oncogene 2013; 32: 4737–4747.
El-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM et al. WAF1, a potential mediator of p53 tumor suppression. Cell 1993; 75: 817–825.
Acknowledgements
We thank Lorenzo Borgognoni, Gianni Gerlini (S. Maria Annunziata Hospital, Florence, Italy), Nicola Pimpinelli (Department of Dermatology, University of Florence, Florence, Italy) and Riccardo Gattai (Department of Medical-Surgical Critical Area, General and Oncological Surgery, University of Florence, Florence, Italy) for providing melanoma samples, Laura Poliseno (Istituto Toscano Tumori, Pisa, Italy) for providing SK-Mel- 2, SK-Mel-5, SK-Mel-28 melanoma cell lines and pCDNA3.1-BRAF wt and pCDNA3.1-BRAF-V600E plasmids, Rosario Notaro (Istituto Toscano Tumori, Florence, Italy) for providing K562 cells, and Silvia Pietrobono for assistance with FACS analysis. This work was supported by a grant from AIRC (Associazione Italiana per la Ricerca sul Cancro IG-14184) to BS.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Riverso, M., Montagnani, V. & Stecca, B. KLF4 is regulated by RAS/RAF/MEK/ERK signaling through E2F1 and promotes melanoma cell growth. Oncogene 36, 3322–3333 (2017). https://doi.org/10.1038/onc.2016.481
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.481
This article is cited by
-
A rapid and stable spontaneous reprogramming system of Spermatogonial stem cells to Pluripotent State
Cell & Bioscience (2023)
-
KLF4 transcription factor in tumorigenesis
Cell Death Discovery (2023)
-
Genome-wide CRISPR knockout screening identified G protein pathway suppressor 2 as a novel tumor suppressor for uveal melanoma metastasis
Journal of Cancer Research and Clinical Oncology (2023)
-
E2F1 rs3213150 polymorphism influences cytarabine sensitivity and prognosis in patients with acute myeloid leukemia
Annals of Hematology (2023)
-
DUB3/KLF4 combats tumor growth and chemoresistance in hepatocellular carcinoma
Cell Death Discovery (2022)