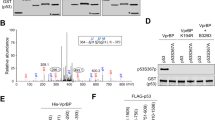
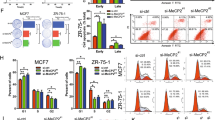
Abstract
p53 is a critical tumor suppressor in humans. It functions mostly as a transcriptional factor and its activity is regulated by numerous post-translational modifications. Among different covalent modifications found on p53 the most controversial one is lysine methylation. We found that human G9a (hG9a) unlike its mouse orthologue (mG9a) potently stimulated p53 transcriptional activity. Both ectopic and endogenous hG9a augmented p53-dependent transcription of pro-apoptotic genes, including Bax and Puma, resulting in enhanced apoptosis and reduced colony formation. Significantly, shRNA-mediated knockdown of hG9a attenuated p53-dependent activation of Puma. On the molecular level, hG9a interacted with histone acetyltransferase, p300/CBP, resulting in increased histone acetylation at the promoter of Puma. The bioinformatics data substantiated our findings showing that positive correlation between G9a and p53 expression is associated with better survival of lung cancer patients. Collectively, this study demonstrates that depending on the cellular and organismal context, orthologous proteins may exert both overlapping and opposing functions. Furthermore, this finding has important ramifications on the use of G9a inhibitors in combination with genotoxic drugs to treat p53-positive tumors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Abbreviations
- DAPI:
-
4,6 diamidino- 2-phenylindole
- GLP:
-
G9a-like protein
- HATs:
-
histone acetyltransferases
- HMTs:
-
histone methyltransferases
- hG9a:
-
human G9a
- hp53:
-
human p53
- KMT:
-
lysine methyltransferase
- mG9a:
-
mouse G9a
- mp53:
-
mouse p53
- PTMs:
-
post-translational modificaitions.
References
Vousden KH, Ryan KM . P53 and Metabolism. Nat Rev Cancer 2009; 9: 691–700.
Kruse J-P, Gu W . Modes of p53 regulation. Cell 2009; 137: 609–622.
Greenberg RA, Chin L, Femino A, Lee K, Gottlieb GJ, Singer RH et al. Short dysfunctional telomeres impair tumorigenesis in the INK4a ⌬ 2/3 cancer-prone mouse. Cell 1999; 97: 515–525.
Stiewe T . The p53 family in differentiation and tumorigenesis. Nat Rev Cancer 2007; 7: 165–168.
Michalak EM, Vandenberg CJ, Delbridge ARD, Wu L, Scott CL, Adams JM et al. Apoptosis-promoted tumorigenesis: gamma-irradiation-induced thymic lymphomagenesis requires Puma-driven leukocyte death. Genes Dev 2010; 24: 1608–1613.
Basu A, Haldar S . The relationship between Bcl2, Bax and p53 : consequences for cell cycle progression and cell death. Mol Hum Reprod 1998; 4: 1099–1109.
Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, Yamashita T et al. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science 2000; 288: 1053–1058.
Hermeking H . p53 enters the microRNA world. Cancer Cell 2007; 12: 414–418.
Lezina L, Purmessur N, Antonov AV, Ivanova T, Karpova E, Krishan K et al. miR-16 and miR-26a target checkpoint kinases Wee1 and Chk1 in response to p53 activation by genotoxic stress. Cell Death Dis 2013; 4: e953.
Zhang A, Xu M, Mo Y . Role of the lncRNA− p53 regulatory network in cancer. J Mol Cell Biol 2014; 6: 181–191.
Pant V, Lozano G . Limiting the power of p53 through the ubiquitin proteasome pathway. Genes Dev 2014; 28: 1739–1751.
Marouco D, Garabadgiu AV, Melino G, Barlev NA . Lysine-specific modifications of p53: a matter of life and death? Oncotarget 2013; 4: 1556–1571.
Moiseeva TN, Bottrill A, Melino G, Barlev NA . DNA damage-induced ubiquitylation of proteasome controls its proteolytic activity. Oncotarget 2013; 44: 1338–1348.
Fedorova O, Moiseeva T, Nikiforov A, Tsimokha A, Livinskaya V, Hodson M et al. Proteomic analysis of the 20 S proteasome (PSMA3)-interacting proteins reveals a functional link between the proteasome and mRNA metabolism. Biochem Biophys Res Commun 2011; 416: 258–265.
Sims RJ, Reinberg D . Is there a code embedded in proteins that is based on post-translational modifications? Nat Rev Mol Cell Biol 2008; 9: 815–820.
Brooks CL, Gu W . p53 Regulation by Ubiquitin. FEBS Lett 2011; 585: 2803–2809.
Espinosa JM, Emerson BM . Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Mol Cell 2001; 8: 57–69.
Barlev NA, Liu L, Chehab NH, Mansfield K, Harris KG, Halazonetis TD et al. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol Cell 2001; 8: 1243–1254.
Ivanov GS, Ivanova T, Kurash J, Ivanov A, Chuikov S, Gizatullin F et al. Methylation-Acetylation interplay activates p53 in response to DNA damage. Mol Cell Biol 2007; 27: 6756–6769.
Liu X, Wang D, Zhao Y, Tu B, Zheng Z, Wang L et al. Methyltransferase Set7/9 regulates p53 activity by interacting with Sirtuin 1 (SIRT1). Proc Natl Acad Sci USA 2011; 108: 1925–1930.
Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA et al. Repression of p53 activity by Smyd2-mediated methylation. Nature 2006; 444: 629–632.
Shi X, Kachirskaia I, Yamaguchi H, West LE, Wen H, Wang EW et al. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol Cell 2007; 27: 636–646.
Huang J, Dorsey J, Chuikov S, Pérez-Burgos L, Zhang X, Jenuwein T et al. G9a and Glp methylate lysine 373 in the tumor suppressor p53. J Biol Chem 2010; 285: 9636–9641.
Chang Y, Zhang X, Horton JR, Upadhyay AK, Spannhoff A, Liu J et al. Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294. Nat Struct Mol Biol 2009; 16: 312–317.
Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H et al. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev 2005; 19: 815–826.
Tachibana M, Sugimoto K, Fukushima T, Shinkai Y . Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem 2001; 276: 25309–25317.
Shinkai Y, Tachibana M . H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev 2011; 25: 781–788.
Kubicek S, O’Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a Histone Methyltransferase. Mol Cell 2007; 25: 473–481.
Trojer P, Zhang J, Yonezawa M, Schmidt A, Zheng H, Jenuwein T et al. Dynamic Histone H1 Isotype 4 Methylation and Demethylation by Histone Lysine Methyltransferase G9a/KMT1C and the Jumonji Domain-containing JMJD2/KDM4 Proteins. J Biol Chem 2009; 284: 8395–8405.
Weiss T, Hergeth S, Zeissler U, Izzo A, Tropberger P, Zee BM et al. Histone H1 variant-specific lysine methylation by G9a/KMT1C and Glp1/KMT1D. Epigenetics Chromatin 2010; 3: 7.
Yu Y, Song C, Zhang Q, DiMaggio PA, Garcia BA, York A et al. Histone H3 Lysine 56 Methylation regulates DNA replication through its interaction with PCNA. Mol Cell 2012; 46: 7–17.
Shankar SR, Bahirvani AG, Rao VK, Bharathy N, Ow JR, Taneja R . G9a, a multipotent regulator of gene expression. Epigenetics 2013; 8: 16–22.
Bittencourt D, Wu D-Y, Jeong KW, Gerke DS, Herviou L, Ianculescu I et al. G9a functions as a molecular scaffold for assembly of transcriptional coactivators on a subset of glucocorticoid receptor target genes. Proc Natl Acad Sci USA 2012; 109: 19673–19678.
Purcell DJ, Jeong KW, Bittencourt D, Gerke DS, Stallcup MR . A distinct mechanism for coactivator versus corepressor function by histone methyltransferase G9a in transcriptional regulation. J Biol Chem 2011; 286: 41963–41971.
Xie P, Tian C, An L, Nie J, Lu K, Xing G et al. Histone methyltransferase protein SETD2 interacts with p53 and selectively regulates its downstream genes. Cell Signal 2008; 20: 1671–1678.
Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS et al. Regulation of p53 activity through lysine methylation. Nature 2004; 432: 353–360.
Brown S, Campbell R, Sanderson C . Novel NG36/G9a gene products encoded within the human and mouse MHC class III regions. Mamm Genome 2001; 12: 916–924.
Collins R, Cheng X . A case study in cross-talk: the histone lysine methyltransferases G9a and GLP. Nucleic Acids Res 2010; 38: 3503–3511.
Sullivan K, Gallant-Behm C, Henry R, Fraikin J, Espinosa J . The p53 circuit board. Biochim Biophys Acta 2012; 1825: 229–244.
Lee DY, Northrop JP, Kuo M-H, Stallcup MR . Histone H3 lysine 9 methyltransferase G9a is a transcriptional coactivator for nuclear receptors. J Biol Chem 2006; 281: 8476–8485.
Gu W, Shi XL, Roeder RG . Synergistic activation of transcription by CBP and p53. Nature 1997; 387: 819–823.
Henry RA, Kuo YM, Andrews AJ . Differences in specificity and selectivity between CBP and p300 acetylation of histone H3 and H3/H4. Biochemistry 2013; 52: 5746–5759.
Antonov AV, Krestyaninova M, Knight R, Rodchenkov I, Melino G, Barlev NA . PPISURV: a novel bioinformatics tool for uncovering the hidden role of specific genes in cancer survival outcome. Oncogene 2014; 33: 1621–1628.
Antonov AV . BioProfiling.de: analytical web portal for high-throughput cell biology. Nucleic Acids Res 2011; 39: W323–W327.
Campaner S, Spreafico F, Burgold T, Doni M, Rosato U, Amati B et al. The Methyltransferase Set7/9 (Setd7) is dispensable for the p53-Mediated DNA damage response in vivo. Mol Cell 2011; 43: 681–688.
Lehnertz B, Rogalski JC, Schulze FM, Yi L, Lin S, Kast J et al. p53-Dependent transcription and tumor suppression are not affected in Set7/9-Deficient Mice. Mol Cell 2011; 43: 673–680.
Chaturvedi C-P, Somasundaram B, Singh K, Carpenedo RL, Stanford WL, Dilworth FJ et al. Maintenance of gene silencing by the coordinate action of the H3K9 methyltransferase G9a/KMT1C and the H3K4 demethylase Jarid1a/KDM5A. Proc Natl Acad Sci USA 2012; 109: 18845–18850.
Dong C, Yuan T, Wu Y, Wang Y, Fan TWM, Miriyala S et al. Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer Cell 2012; 23: 316–331.
Mulligan P, Westbrook TF, Ottinger M, Pavlova N, Chang B, Macia E et al. CDYL bridges REST and Histone Methyltransferases for Gene repression and suppression of cellular transformation. Mol Cell 2008; 32: 718–726.
Oh ST, Kim KB, Chae YC, Kang JY, Hahn Y, Seo SB . H3K9 histone methyltransferase G9a-mediated transcriptional activation of p21. FEBS Lett 2014; 588: 685–691.
Thandapani P, O’Connor TR, Bailey TL, Richard S . Defining the RGG/RG Motif. Mol Cell 2013; 50: 613–623.
Lezina L, Aksenova V, Ivanova T, Purmessur N, Antonov AV, Tentler D et al. KMTase Set7/9 is a critical regulator of E2F1 activity upon genotoxic stress. Cell Death Differ 2014; 21: 1889–1899.
Lezina L, Aksenova V, Fedorova O, Malikova D, Shuvalov O, Antonov AV et al. KMT Set7/9 affects genotoxic stress response via the Mdm2 axis. Oncotarget 2015; 6: 25843–25855.
Celardo I, Grespi F, Antonov A, Bernassola F, Garabadgiu AV, Melino G et al. Caspase-1 is a novel target of p63 in tumor suppression. Cell Death Dis 2013; 4: e645.
Acknowledgements
We thank Dr O. Binda (Newcastle University, UK), Dr MR. Stallcup (University of Southern California, USA), Dr C-M Chiang (University of Texas Southwestern Medical Center, USA), Dr SB Seo (Chung-Ang University, Korea), Dr P. Muller (MRC Toxicology Unit, UK) for providing various constructs and reagents. We also thank S Cowley lab (University of Leicester, UK) for providing E14 cells. This work was supported by grants from RSF 14-15-00816 to (E.V. and N.B.); M.R. was supported by a Doctoral Studentship from the Kurdistan Regional Government (Iraq); A.V.A. was supported by MCB RAS programme; G.M. acknowledges the grant support from Medical Research Council, AIRC 2014 IG15653, AIRC 5xmille MCO9979, AIRC 2011 IG11955 and Fondazione Roma malattie Non trasmissibili Cronico-Degenerative (NCD).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Rada, M., Vasileva, E., Lezina, L. et al. Human EHMT2/G9a activates p53 through methylation-independent mechanism. Oncogene 36, 922–932 (2017). https://doi.org/10.1038/onc.2016.258
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.258
This article is cited by
-
The novel indomethacin derivative CZ-212-3 exerts antitumor effects on castration-resistant prostate cancer by degrading androgen receptor and its variants
Acta Pharmacologica Sinica (2022)
-
Runt related transcription factor-1 plays a central role in vessel co-option of colorectal cancer liver metastases
Communications Biology (2021)
-
SETD3 is a positive regulator of DNA-damage-induced apoptosis
Cell Death & Disease (2019)
-
Inhibition of G9a by a small molecule inhibitor, UNC0642, induces apoptosis of human bladder cancer cells
Acta Pharmacologica Sinica (2019)