Abstract
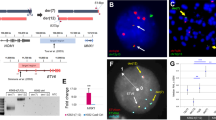
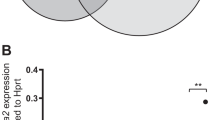
Acute myeloid leukemia (AML) is a heterogeneous disease comprising a large number of subtypes defined by specific chromosome abnormalities. One such subtype carries the t(6;9)(p22;q34) chromosome rearrangement, which leads to expression of the DEK-NUP214 chimeric gene, and has a particularly poor outcome. To provide a better understanding of the molecular etiology of these relatively rare individual AML variants, it is necessary to generate in vivo models, which can also serve as a means to evaluate targeted therapies based on their specific genetic abnormalities. Here, we describe the development of a human cell AML, generated in CD34+ human hematopoietic progenitor cells xenografted into immunocompromised mice that express human myeloid cell growth factors. Within 6 months, these mice develop a human cell AML with phenotypic characteristics of the primary t(6;9) disease and a CD45+CD13+CD34+CD38+ immunophenotype. Gene expression studies show that members of the HOX family of genes (HOXA9, 10, B3, B4 and PBX3) are highly upregulated in the AML from this mouse model as well as from primary human t(6;9) AML. Gene expression analysis also identified several other significantly disregulated pathways involving KRAS, BRCA1 and ALK, for example. This is the first report of a humanized model of the DEK-NUP214 disease and provides a means to study the development and treatment of this particular subtype of AML.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Soekarman D, von Lindern M, Daenen S, de Jong B, Fonatsch C, Heinze B et al. The translocation (6;9) (p23;q34) shows consistent rearrangement of two genes and defines a myeloproliferative disorder with specific clinical features. Blood 1992; 79: 2990–2997.
von Lindern M, Fornerod M, van Baal S, Jaegle M, de Wit T, Buijs et al. The translocation (6;9), associated with a specific subtype of acute myeloid leukemia, results in the fusion of two genes, dek and can, and the expression of a chimeric, leukemia-specific dek-can mRNA. Mol Cell Biol 1992; 12: 1687–1697.
Slovak ML, Gundacker H, Bloomfield CD, Dewald G, Appelbaum FR, Larson RA et al. A retrospective study of 69 patients with t(6;9)(p23;q34) AML emphasizes the need for a prospective, multicenter initiative for rare 'poor prognosis' myeloid malignancies. Leukemia 2006; 20: 1295–1297.
Oyarzo MP, Lin P, Glassman A, Bueso-Ramos CE, Luthra R, Medeiros LJ . Acute myeloid leukemia with t(6;9)(p23;q34) is associated with dysplasia and a high frequency of flt3 gene mutations. Am J Clin Pathol 2004; 122: 348–358.
Sanden C, Gullberg U . The DEK oncoprotein and its emerging roles in gene regulation. Leukemia 2015; 29: 1632–1636.
Riveiro-Falkenbach E, Soengas MS . Control of tumorigenesis and chemoresistance by the DEK oncogene. Clin Cancer Res 2010; 16: 2932–2938.
Privette Vinnedge LM, Kappes F, Nassar N, Wells SI . Stacking the DEK: from chromatin topology to cancer stem cells. Cell Cycle 2013; 12: 51–66.
Privette Vinnedge LM, Ho SM, Wikenheiser-Brokamp KA, Wells SI . The DEK oncogene is a target of steroid hormone receptor signaling in breast cancer. PLoS One 2012; 7: e46985.
Wise-Draper TM, Mintz-Cole RA, Morris TA, Simpson DS, Wikenheiser-Brokamp KA, Currier MA et al. Overexpression of the cellular DEK protein promotes epithelial transformation in vitro and in vivo. Cancer Res 2009; 69: 1792–1799.
Ageberg M, Gullberg U, Lindmark A . The involvement of cellular proliferation status in the expression of the human proto-oncogene DEK. Haematologica 2006; 91: 268–269.
Kraemer D, Wozniak RW, Blobel G, Radu A . The human CAN protein, a putative oncogene product associated with myeloid leukemogenesis, is a nuclear pore complex protein that faces the cytoplasm. Proc Natl Acad Sci USA 1994; 91: 1519–1523.
Sanden C, Ageberg M, Petersson J, Lennartsson A, Gullberg U . Forced expression of the DEK-NUP214 fusion protein promotes proliferation dependent on upregulation of mTOR. BMC Cancer 2013; 13: 440.
Oancea C, Ruster B, Henschler R, Puccetti E, Ruthardt M . The t(6;9) associated DEK/CAN fusion protein targets a population of long-term repopulating hematopoietic stem cells for leukemogenic transformation. Leukemia 2010; 24: 1910–1919.
Palacios R, Henson G, Steinmetz M, McKearn JP . Interleukin-3 supports growth of mouse pre-B-cell clones in vitro. Nature 1984; 309: 126–131.
Avanzi GC, Brizzi MF, Giannotti J, Ciarletta A, Yang YC, Pegoraro L et al. M-07e human leukemic factor-dependent cell line provides a rapid and sensitive bioassay for the human cytokines GM-CSF and IL-3. J Cell Physiol 1990; 145: 458–464.
Ren M, Qin H, Kitamura E, Cowell JK . Dysregulated signaling pathways in the development of CNTRL-FGFR1-induced myeloid and lymphoid malignancies associated with FGFR1 in human and mouse models. Blood 2013; 122: 1007–1016.
Ren M, Qin H, Ren R, Cowell JK . Ponatinib suppresses the development of myeloid and lymphoid malignancies associated with FGFR1 abnormalities. Leukemia 2013; 27: 32–40.
Billerbeck E, Barry WT, Mu K, Dorner M, Rice CM, Ploss A . Development of human CD4+FoxP3+ regulatory T cells in human stem cell factor-, granulocyte-macrophage colony-stimulating factor-, and interleukin-3-expressing NOD-SCID IL2Rgamma(null) humanized mice. Blood 2011; 117: 3076–3086.
Yamauchi K, Yasuda M . Comparison in treatments of nonleukemic granulocytic sarcoma: report of two cases and a review of 72 cases in the literature. Cancer 2002; 94: 1739–1746.
Balgobind BV, Van den Heuvel-Eibrink MM, De Menezes RX, Reinhardt D, Hollink IH, Arentsen-Peters ST et al. Evaluation of gene expression signatures predictive of cytogenetic and molecular subtypes of pediatric acute myeloid leukemia. Haematologica 2011; 96: 221–230.
Mills KI, Kohlmann A, Williams PM, Wieczorek L, Liu WM, Li R et al. Microarray-based classifiers and prognosis models identify subgroups with distinct clinical outcomes and high risk of AML transformation of myelodysplastic syndrome. Blood 2009; 114: 1063–1072.
Saito Y, Nakahata S, Yamakawa N, Kaneda K, Ichihara E, Suekane et al. CD52 as a molecular target for immunotherapy to treat acute myeloid leukemia with high EVI1 expression. Leukemia 2011; 25: 921–931.
Hamaguchi H, Nagata K, Yamamoto K, Fujikawa I, Kobayashi M, Eguchi M . Establishment of a novel human myeloid leukaemia cell line (FKH-1) with t(6;9)(p23;q34) and the expression of dek-can chimaeric transcript. Br J Haematol 1998; 102: 1249–1256.
Ren M, Cowell JK . Constitutive Notch pathway activation in murine ZMYM2-FGFR1-induced T-cell lymphomas associated with atypical myeloproliferative disease. Blood 2011; 117: 6837–6847.
Spencer DH, Young MA, Lamprecht TL, Helton NM, Fulton R, O'Laughlin M et al. Epigenomic analysis of the HOX gene loci reveals mechanisms that may control canonical expression patterns in AML and normal hematopoietic cells. Leukemia 2015; 29: 1279–1289.
Abramovich C, Humphries RK . Hox regulation of normal and leukemic hematopoietic stem cells. Curr Opin Hematol 2005; 12: 210–216.
Alharbi RA, Pettengell R, Pandha HS, Morgan R . The role of HOX genes in normal hematopoiesis and acute leukemia. Leukemia 2013; 27: 1000–1008.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Rights and permissions
About this article
Cite this article
Qin, H., Malek, S., Cowell, J. et al. Transformation of human CD34+ hematopoietic progenitor cells with DEK-NUP214 induces AML in an immunocompromised mouse model. Oncogene 35, 5686–5691 (2016). https://doi.org/10.1038/onc.2016.118
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.118
This article is cited by
-
DEK-AFF2 fusion-associated papillary squamous cell carcinoma of the sinonasal tract: clinicopathologic characterization of seven cases with deceptively bland morphology
Modern Pathology (2021)
-
Acute Myeloid Leukemia in Children: Emerging Paradigms in Genetics and New Approaches to Therapy
Current Oncology Reports (2021)