Abstract
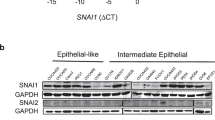
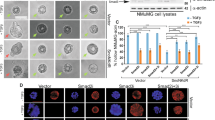
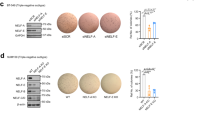
HMG20A is a high mobility group (HMG) domain containing protein homologous to HMG20B, a core subunit of the Lys-specific demethylase 1/REST co-repressor 1 (LSD1-CoREST) histone demethylase complex. Here, we show that HMG20A can replace HMG20B and, therefore, they are mutually exclusive subunits of the complex. Both proteins interact through a coiled-coil domain with BHC80, another subunit of the LSD1-CoREST complex. To investigate the functional differences between the two proteins, we performed transcriptomic analysis of HMG20A- and HMG20B-depleted cells. Analysis of the misregulated genes in HMG20A-knockdown cells evidenced a high proportion of genes related to the epithelial-to-mesenchymal transition (EMT) process. EMT occurs during embryonic development or during the course of malignant cancer progression and consists in the dynamic and reversible transitions between epithelial and mesenchymal phenotypes. We show that HMG20A together with LSD1 are required for SNAI1-dependent repression of epithelial genes and for (transforming growth factor β) TGF-β-triggered EMT. Importantly, HMG20A-depleted cells displayed reduced binding of LSD1 to epithelial gene promoters and increased methylation of lysine 4 of histone H3, suggesting a role of HMG20A in recruiting or in stabilizing the complex at the chromatin. SNAI1 and the TGF-β-related transcription factor SMAD4 were found to be associated with the LSD1-CoREST complex containing HMG20A. Furthermore, we show that HMG20A-depleted cells displayed reduced motility and invasion activity. Finally, we show that expression of HMG20A correlates positively with mesenchymal markers and negatively with epithelial markers in human tumor samples. Taken together, our data demonstrate that HMG20A is essential for the mesenchymal phenotype.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Lamouille S, Xu J, Derynck R . Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 2014; 15: 178–196.
Nieto MA . Epithelial plasticity: a common theme in embryonic and cancer cells. Science 2013; 342: 1234850.
Nieto MA . The ins and outs of the epithelial to mesenchymal transition in health and disease. Annu Rev Cell Dev Biol 2011; 27: 347–376.
Kalluri R, Weinberg RA . The basics of epithelial-mesenchymal transition. J Clin Invest 2009; 119: 1420–1428.
Tsai JH, Yang J . Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev 2013; 27: 2192–2206.
Barrallo-Gimeno A, Nieto MA . The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development 2005; 132: 3151–3161.
Wu CY, Tsai YP, Wu MZ, Teng SC, Wu KJ . Epigenetic reprogramming and post-transcriptional regulation during the epithelial-mesenchymal transition. Trends Genet 2012; 28: 454–463.
Tam WL, Weinberg RA . The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med 2013; 19: 1438–1449.
Lin T, Ponn A, Hu X, Law BK, Lu J . Requirement of the histone demethylase LSD1 in Snai1-mediated transcriptional repression during epithelial-mesenchymal transition. Oncogene 2010; 29: 4896–4904.
Lin Y, Wu Y, Li J, Dong C, Ye X, Chi YI, Evers BM, Zhou BP . The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J 2010; 29: 1803–1816.
Herranz N, Pasini D, Diaz VM, Franci C, Gutierrez A, Dave N et al. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol Cell Biol 2008; 28: 4772–4781.
Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004; 119: 941–953.
Lee MG, Wynder C, Cooch N, Shiekhattar R . An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature 2005; 437: 432–435.
Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 2005; 437: 436–439.
Lim S, Janzer A, Becker A, Zimmer A, Schule R, Buettner R et al. Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis 2010; 31: 512–520.
Ferrari-Amorotti G, Fragliasso V, Esteki R, Prudente Z, Soliera AR, Cattelani S et al. Inhibiting interactions of lysine demethylase LSD1 with snail/slug blocks cancer cell invasion. Cancer Res 2012; 73: 235–245.
McDonald OG, Wu H, Timp W, Doi A, Feinberg AP . Genome-scale epigenetic reprogramming during epithelial-to-mesenchymal transition. Nat Struct Mol Biol 2011; 18: 867–874.
Saleque S, Kim J, Rooke HM, Orkin SH . Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Mol Cell 2007; 27: 562–572.
Shi Y, Sawada J, Sui G, Affar el B, Whetstine JR, Lan F et al. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 2003; 422: 735–738.
Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y . Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell 2005; 19: 857–864.
Hakimi MA, Bochar DA, Chenoweth J, Lane WS, Mandel G, Shiekhattar R . A core-BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc Natl Acad Sci USA 2002; 99: 7420–7425.
You A, Tong JK, Grozinger CM, Schreiber SL . CoREST is an integral component of the CoREST- human histone deacetylase complex. Proc Natl Acad Sci USA 2001; 98: 1454–1458.
Marmorstein LY, Kinev AV, Chan GK, Bochar DA, Beniya H, Epstein JA et al. A human BRCA2 complex containing a structural DNA binding component influences cell cycle progression. Cell 2001; 104: 247–257.
Wynder C, Hakimi MA, Epstein JA, Shilatifard A, Shiekhattar R . Recruitment of MLL by HMG-domain protein iBRAF promotes neural differentiation. Nat Cell Biol 2005; 7: 1113–1117.
Ceballos-Chavez M, Rivero S, Garcia-Gutierrez P, Rodriguez-Paredes M, Garcia-Dominguez M, Bhattacharya S et al. Control of neuronal differentiation by sumoylation of BRAF35, a subunit of the LSD1-CoREST histone demethylase complex. Proc Natl Acad Sci USA 2012.
Iwase S, Januma A, Miyamoto K, Shono N, Honda A, Yanagisawa J . Characterization of BHC80 in BRAF-HDAC complex, involved in neuron-specific gene repression. Biochem Biophys Res Commun 2004; 322: 601–608.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550.
Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, Yang J et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci USA 2010; 107: 15449–15454.
Barbera MJ, Puig I, Dominguez D, Julien-Grille S, Guaita-Esteruelas S, Peiro S et al. Regulation of Snail transcription during epithelial to mesenchymal transition of tumor cells. Oncogene 2004; 23: 7345–7354.
McKay BS, Irving PE, Skumatz CM, Burke JM . Cell-cell adhesion molecules and the development of an epithelial phenotype in cultured human retinal pigment epithelial cells. Exp Eye Res 1997; 65: 661–671.
Miettinen PJ, Ebner R, Lopez AR, Derynck R . TGF-beta induced transdifferentiation of mammary epithelial cells to mesenchymal cells: involvement of type I receptors. J Cell Biol 1994; 127: 2021–2036.
Massague J . TGFbeta signalling in context. Nat Rev Mol Cell Biol 2012; 13: 616–630.
Li H, Wang H, Wang F, Gu Q, Xu X . Snail involves in the transforming growth factor beta1-mediated epithelial-mesenchymal transition of retinal pigment epithelial cells. PLoS One 2011; 6: e23322.
Xu J, Lamouille S, Derynck R . TGF-beta-induced epithelial to mesenchymal transition. Cell Res 2009; 19: 156–172.
Vincent T, Neve EP, Johnson JR, Kukalev A, Rojo F, Albanell J et al. A SNAIL1-SMAD3/4 transcriptional repressor complex promotes TGF-beta mediated epithelial-mesenchymal transition. Nat Cell Biol 2009; 11: 943–950.
Yilmaz M, Christofori G . EMT the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev 2009; 28: 15–33.
Yilmaz M, Christofori G . Mechanisms of motility in metastasizing cells. Mol Cancer Res 2010; 8: 629–642.
Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D . ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 2004; 6: 1–6.
Xu L, Shen SS, Hoshida Y, Subramanian A, Ross K, Brunet JP et al. Gene expression changes in an animal melanoma model correlate with aggressiveness of human melanoma metastases. Mol Cancer Res 2008; 6: 760–769.
Talantov D, Mazumder A, Yu JX, Briggs T, Jiang Y, Backus J et al. Novel genes associated with malignant melanoma but not benign melanocytic lesions. Clin Cancer Res 2005; 11: 7234–7242.
Riker AI, Enkemann SA, Fodstad O, Liu S, Ren S, Morris C et al. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med Genomics 2008; 1: 13.
Gordon GJ, Rockwell GN, Jensen RV, Rheinwald JG, Glickman JN, Aronson JP et al. Identification of novel candidate oncogenes and tumor suppressors in malignant pleural mesothelioma using large-scale transcriptional profiling. Am J Pathol 2005; 166: 1827–1840.
Barretina J, Taylor BS, Banerji S, Ramos AH, Lagos-Quintana M, Decarolis PL et al. Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nat Genet 2010; 42: 715–721.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013; 6: pl1.
Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet 2012; 44: 685–689.
Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013; 499: 43–49.
Wang Y, Zhang H, Chen Y, Sun Y, Yang F, Yu W et al. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell 2009; 138: 660–672.
Onder TT, Gupta PB, Mani SA, Yang J, Lander ES, Weinberg RA . Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res 2008; 68: 3645–3654.
Radisky DC, Levy DD, Littlepage LE, Liu H, Nelson CM, Fata JE et al. Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature 2005; 436: 123–127.
Colland F, Jacq X, Trouplin V, Mougin C, Groizeleau C, Hamburger A et al. Functional proteomics mapping of a human signaling pathway. Genome Res 2004; 14: 1324–1332.
Dong C, Wu Y, Yao J, Wang Y, Yu Y, Rychahou PG et al. G9a interacts with Snail and is critical for Snail-mediated E-cadherin repression in human breast cancer. J Clin Invest 2012; 122: 1469–1486.
Millanes-Romero A, Herranz N, Perrera V, Iturbide A, Loubat-Casanovas J, Gil J et al. Regulation of heterochromatin transcription by Snail1/LOXL2 during epithelial-to-mesenchymal transition. Mol Cell 2013; 52: 746–757.
Sumoy L, Carim L, Escarceller M, Nadal M, Gratacos M, Pujana MA et al. HMG20A and HMG20B map to human chromosomes 15q24 and 19p13.3 and constitute a distinct class of HMG-box genes with ubiquitous expression. Cytogenet Cell Genet 2000; 88: 62–67.
Subtil-Rodriguez A, Reyes JC . BRG1 helps RNA polymerase II to overcome a nucleosomal barrier during elongation, in vivo. EMBO Rep 2010; 11: 751–757.
Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP . Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 2003; 31: e15.
Sanges R, Cordero F, Calogero RA . oneChannelGUI: a graphical interface to Bioconductor tools, designed for life scientists who are not familiar with R language. Bioinformatics 2007; 23: 3406–3408.
Huang da W, Sherman BT, Lempicki RA . Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4: 44–57.
Liang CC, Park AY, Guan JL . In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc 2007; 2: 329–333.
Hazan RB, Phillips GR, Qiao RF, Norton L, Aaronson SA . Exogenous expression of N-cadherin in breast cancer cells induces cell migration, invasion, and metastasis. J Cell Biol 2000; 148: 779–790.
Acknowledgements
We thank L Sumoy, T Baba, A García de Herreros and J Lu for reagents and plasmids. We thank Dr M Garcia-Dominguez and Dr JA Pintor-Toro for critical reading of the manuscript. We thank E Andújar and M Pérez from the CABIMER Genomic Unit for microarray expression analysis. The results published here are in part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/. This work was funded by the Spanish Ministry of Economy and Competitiveness [BFU-2011-23442 to JCR] and a Juan de la Cierva grant to SR and by Junta de Andalucía (P09-CTS-04967 to SSB).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Rivero, S., Ceballos-Chávez, M., Bhattacharya, S. et al. HMG20A is required for SNAI1-mediated epithelial to mesenchymal transition. Oncogene 34, 5264–5276 (2015). https://doi.org/10.1038/onc.2014.446
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2014.446
This article is cited by
-
Pan-cancer onco-signatures reveal a novel mitochondrial subtype of luminal breast cancer with specific regulators
Journal of Translational Medicine (2023)
-
A developmental role for the chromatin-regulating CoREST complex in the cnidarian Nematostella vectensis
BMC Biology (2022)
-
HMG20A was identified as a key enhancer driver associated with DNA damage repair in oral squamous cell carcinomas
BMC Oral Health (2022)
-
SFMBT1 facilitates colon cancer cell metastasis and drug resistance combined with HMG20A
Cell Death Discovery (2022)
-
HMG20B stabilizes association of LSD1 with GFI1 on chromatin to confer transcription repression and leukemia cell differentiation block
Oncogene (2022)