Abstract
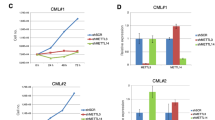
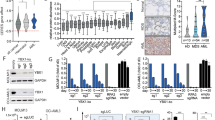
High activation of the PI3K–AKT–mTOR pathway is characteristic for T-cell acute lymphoblastic leukemia (T-ALL). The activity of the master regulator of this pathway, PTEN, is often impaired in T-ALL. However, experimental evidence suggests that input from receptor tyrosine kinases (RTKs) is required for sustained mTOR activation, even in the absence of PTEN. We previously reported the expression of Neurotrophin receptor tyrosine kinases (TRKs) and their respective ligands in primary human leukemia samples. In the present study we aimed to dissect the downstream signaling cascades of TRK-induced T-ALL in a murine model and show that T-ALLs induced by deregulated receptor tyrosine kinase signaling acquire activating mutations in Notch1 and lose PTEN during clonal evolution. Some clones additionally lost one allele of the homeodomain transcription factor Cux1. All events independently led to a gradual hyperactivation of both mTORC1 and mTORC2 signaling. We dissected the role of the individual mTOR complexes by shRNA knockdown and found that the separate depletion of mTORC1 or mTORC2 reduced the growth of T-ALL blasts, but was not sufficient to induce apoptosis. In contrast, knockdown of the mTOR downstream effector eIF4E caused a striking cytotoxic effect, demonstrating a critical addiction to cap-dependent mRNA-translation. Although high mTORC2–AKT activation is commonly associated with drug-resistance, we demonstrate that T-ALL displaying a strong mTORC2–AKT activation were specifically susceptible to 4EGI-1, an inhibitor of the eIF4E–eIF4G interaction. To decipher the mechanism of 4EGI-1, we performed a genome-wide analysis of mRNAs that are translationally regulated by 4EGI-1 in T-ALL. 4EGI-1 effectively reduced the ribosomal occupancy of mRNAs that were strongly upregulated in T-ALL blasts compared with normal thymocytes including transcripts important for translation, mitochondria and cell cycle progression, such as cyclins and ribosomal proteins. These data suggest that disrupting the eIF4E–eIF4G interaction constitutes a promising therapy strategy in mTOR-deregulated T-cell leukemia.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Gokbuget N, Stanze D, Beck J, Diedrich H, Horst HA, Huttmann A et al. Outcome of relapsed adult lymphoblastic leukemia depends on response to salvage chemotherapy, prognostic factors, and performance of stem cell transplantation. Blood 2012; 120: 2032–2041.
Van Vlierberghe P, Pieters R, Beverloo HB, Meijerink JP . Molecular-genetic insights in paediatric T-cell acute lymphoblastic leukaemia. Br J Haematol 2008; 143: 153–168.
Homminga I, Pieters R, Langerak AW, de Rooi JJ, Stubbs A, Verstegen M et al. Integrated transcript and genome analyses reveal NKX2-1 and MEF2C as potential oncogenes in T cell acute lymphoblastic leukemia. Cancer Cell 2011; 19: 484–497.
Weng AP, Ferrando AA, Lee W, Morris JP, Silverman LB, Sanchez-Irizarry C et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 2004; 306: 269–271.
Aster JC, Pear WS, Blacklow SC . Notch Signaling in Leukemia. Annu Rev Pathol 2008; 3: 587–613.
Ntziachristos P, Tsirigos A, Van Vlierberghe P, Nedjic J, Trimarchi T, Flaherty MS et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat Med 2012; 18: 298–301.
Chiang MY, Xu L, Shestova O, Histen G, L’heureux S, Romany C et al. Leukemia-associated NOTCH1 alleles are weak tumor initiators but accelerate K-ras–initiated leukemia. J Clin Invest 2008; 118: 3181–3194.
Chan SM, Weng AP, Tibshirani R, Aster JC, Utz PJ . Notch signals positively regulate activity of the mTOR pathway in T-cell acute lymphoblastic leukemia. Blood 2007; 110: 278–286.
Silva A, Yunes JA, Cardoso BA, Martins LR, Jotta PY, Abecasis M et al. PTEN posttranslational inactivation and hyperactivation of the PI3K/Akt pathway sustain primary T cell leukemia viability. J Clin Invest 2008; 118: 3762–3774.
Palomero T, Sulis ML, Cortina M, Real PJ, Barnes K, Ciofani M et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med 2007; 13: 1203–1210.
Clappier E, Gerby B, Sigaux F, Delord M, Touzri F, Hernandez L et al. Clonal selection in xenografted human T cell acute lymphoblastic leukemia recapitulates gain of malignancy at relapse. J Exp Med 2011; 208: 653–661.
Wei G, Twomey D, Lamb J, Schlis K, Agarwal J, Stam RW et al. Gene expression-based chemical genomics identifies rapamycin as a modulator of MCL1 and glucocorticoid resistance. Cancer Cell 2006; 10: 331–342.
Piovan E, Yu J, Tosello V, Herranz D, Ambesi-Impiombato A, Da Silva AC et al. Direct reversal of glucocorticoid resistance by AKT inhibition in acute lymphoblastic leukemia. Cancer Cell 2013; 24: 766–776.
Blackburn JS, Liu S, Wilder JL, Dobrinski KP, Lobbardi R, Moore FE et al. Clonal evolution enhances leukemia-propagating cell frequency in T cell acute lymphoblastic leukemia through Akt/mTORC1 pathway activation. Cancer Cell 2014; 25: 366–378.
Gutierrez A, Sanda T, Grebliunaite R, Carracedo A, Salmena L, Ahn Y et al. High frequency of PTEN, PI3K, and AKT abnormalities in T-cell acute lymphoblastic leukemia. Blood 2009; 114: 647–650.
Trinquand A, Tanguy-Schmidt A, Ben Abdelali R, Lambert J, Beldjord K, Lengline E et al. Toward a NOTCH1/FBXW7/RAS/PTEN-based oncogenetic risk classification of adult T-cell acute lymphoblastic leukemia: a Group for Research in Adult Acute Lymphoblastic Leukemia study. J Clin Oncol 2013; 31: 4333–4342.
Gutierrez A, Grebliunaite R, Feng H, Kozakewich E, Zhu S, Guo F et al. Pten mediates Myc oncogene dependence in a conditional zebrafish model of T cell acute lymphoblastic leukemia. J Exp Med 2011; 208: 1595–1603.
Bonnet M, Loosveld M, Montpellier B, Navarro JM, Quilichini B, Picard C et al. Posttranscriptional deregulation of MYC via PTEN constitutes a major alternative pathway of MYC activation in T-cell acute lymphoblastic leukemia. Blood 2011; 117: 6650–6659.
Subramaniam PS, Whye DW, Efimenko E, Chen J, Tosello V, De Keersmaecker K et al. Targeting nonclassical oncogenes for therapy in T-ALL. Cancer Cell 2012; 21: 459–472.
Curry NL, Mino-Kenudson M, Oliver TG, Yilmaz OH, Yilmaz VO, Moon JY et al. Pten-null tumors cohabiting the same lung display differential AKT activation and sensitivity to dietary restriction. Cancer Discov 2013; 3: 908–921.
Ren M, Li X, Cowell JK . Genetic fingerprinting of the development and progression of T-cell lymphoma in a murine model of atypical myeloproliferative disorder initiated by the ZNF198-fibroblast growth factor receptor-1 chimeric tyrosine kinase. Blood 2009; 114: 1576–1584.
Wasag B, Lierman E, Meeus P, Cools J, Vandenberghe P . The kinase inhibitor TKI258 is active against the novel CUX1-FGFR1 fusion detected in a patient with T-lymphoblastic leukemia/lymphoma and t(7;8)(q22;p11). Haematologica 2011; 96: 922–926.
Li Z, Beutel G, Rhein M, Meyer J, Koenecke C, Neumann T et al. High-affinity neurotrophin receptors and ligands promote leukemogenesis. Blood 2009; 113: 2028–2037.
Segal RA . Selectivity in neurotrophin signaling: theme and variations. Annu Rev Neurosci 2003; 26: 299–330.
Mulloy JC, Jankovic V, Wunderlich M, Delwel R, Cammenga J, Krejci O et al. AML1-ETO fusion protein up-regulates TRKA mRNA expression in human CD34+ cells, allowing nerve growth factor-induced expansion. Proc Natl Acad Sci USA 2005; 102: 4016–4021.
Pearse RN . A neurotrophin axis in myeloma: TrkB and BDNF promote tumor-cell survival. Blood 2005; 105: 4429–4436.
Renné C, Minner S, Küppers R, Hansmann M-L, Bräuninger A . Autocrine NGFbeta/TRKA signalling is an important survival factor for Hodgkin lymphoma derived cell lines. Leuk Res 2008; 32: 163–167.
Graeber TG, Eisenberg D . Eisenberg_Bioinformatic identification of autocrine loops. Nat Genet 2001; 29: 295–300.
Meyer J, Rhein M, Schiedlmeier B, Kustikova O, Rudolph C, Kamino K et al. Remarkable leukemogenic potency and quality of a constitutively active neurotrophin receptor, ΔTrkA. Leukemia 2007; 21: 2171–2180.
Soulier J . HOXA genes are included in genetic and biologic networks defining human acute T-cell leukemia (T-ALL). Blood 2005; 106: 274–286.
Lee JO, Yang H, Georgescu MM, Di Cristofano A, Maehama T, Shi Y et al. Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell 1999; 99: 323–334.
Ashworth TD, Pear WS, Chiang MY, Blacklow SC, Mastio J, Xu L et al. Deletion-based mechanisms of Notch1 activation in T-ALL: key roles for RAG recombinase and a conserved internal translational start site in Notch1. Blood 2010; 116: 5455–5464.
Kleppe M, Mentens N, Tousseyn T, Wlodarska I, Cools J . MOHITO, a novel mouse cytokine-dependent T-cell line, enables studies of oncogenic signaling in the T-cell context. Haematologica 2011; 96: 779–783.
Wong CC, Martincorena I, Rust AG, Rashid M, Alifrangis C, Alexandrov LB et al. Inactivating CUX1 mutations promote tumorigenesis. Nat Genet 2013; 46: 33–38.
Fellmann C, Hoffmann T, Sridhar V, Hopfgartner B, Muhar M, Roth M et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep 2013; 5: 1704–1713.
Rhein M, Schwarzer A, Yang M, Kaever V, Brugman M, Meyer J et al. Leukemias induced by altered TRK-signaling are sensitive to mTOR inhibitors in preclinical models. Ann Hematol 2011; 90: 283–292.
Moerke NJ, Aktas H, Chen H, Cantel S, Reibarkh MY, Fahmy A et al. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell 2007; 128: 257–267.
Chen L, Aktas BH, Wang Y, He X, Sahoo R, Zhang N et al. Tumor suppression by small molecule inhibitors of translation initiation. Oncotarget 2012; 3: 869–881.
Hsieh AC, Costa M, Zollo O, Davis C, Feldman ME, Testa JR et al. Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP-eIF4E. Cancer Cell 2010; 17: 249–261.
Schatz JH, Oricchio E, Wolfe AL, Jiang M, Linkov I, Maragulia J et al. Targeting cap-dependent translation blocks converging survival signals by AKT and PIM kinases in lymphoma. J Exp Med 2011; 208: 1799–1807.
Tamburini J, Green AS, Bardet V, Chapuis N, Park S, Willems L et al. Protein synthesis is resistant to rapamycin and constitutes a promising therapeutic target in acute myeloid leukemia. Blood 2009; 114: 1618–1627.
Lin C-J, Nasr Z, Premsrirut PK, Porco J, John A, Hippo Y, Lowe SW et al. Targeting synthetic lethal interactions between Myc and the eIF4F complex impedes tumorigenesis. Cell Rep 2012; 1: 325–333.
De Benedetti A, Graff JR . eIF-4E expression and its role in malignancies and metastases. Oncogene 2004; 23: 3189–3199.
Ramsay G, Evan GI, Bishop JM . The protein encoded by the human proto-oncogene c-myc. Proc Natl Acad Sci USA 1984; 81: 7742–7746.
Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM . A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 2012; 486: 109–113.
Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 2012; 485: 55–61.
Laplante M, Sabatini DM . mTOR signaling in growth control and disease. Cell 2012; 149: 274–293.
Lee K, Nam KT, Cho SH, Gudapati P, Hwang Y, Park D-S et al. Vital roles of mTOR complex 2 in Notch-driven thymocyte differentiation and leukemia. J Exp Med 2012; 209: 713–728.
Fu Z, Tindall DJ . FOXOs, cancer and regulation of apoptosis. Oncogene 2008; 27: 2312–2319.
Jo OD, Martin J, Bernath A, Masri J, Lichtenstein A, Gera J . Heterogeneous nuclear ribonucleoprotein A1 regulates cyclin D1 and c-myc internal ribosome entry site function through Akt signaling. J Biol Chem 2008; 283: 23274–23287.
Birsoy K, Possemato R, Lorbeer FK, Bayraktar EC, Thiru P, Yucel B et al. Metabolic determinants of cancer cell sensitivity to glucose limitation and biguanides. Nature 2014; 508: 108–112.
Dello Sbarba P, Rovida E, Marzi I, Cipolleschi MG . One more stem cell niche: how the sensitivity of chronic myeloid leukemia cells to imatinib mesylate is modulated within a ‘hypoxic’ environment. Hypoxia 2014; 2: 1–10.
Lagadinou ED, Sach A, Callahan K, Rossi RM, Neering SJ, Minhajuddin M et al. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell 2013; 12: 329–341.
Lee JY, Nakada D, Yilmaz ÖH, Tothova Z, Joseph NM, Lim MS et al. mTOR activation induces tumor suppressors that inhibit leukemogenesis and deplete hematopoietic stem cells after Pten deletion. Cell Stem Cell 2010; 7: 593–605.
Larsson O, Sonenberg N, Nadon R . anota: analysis of differential translation in genome-wide studies. Bioinformatics 2011; 27: 1440–1441.
Acknowledgements
AS was supported by the Hannover Biomedical Research School (HBRS) and the MD-PhD program ‘Molecular Medicine’. We thank Dr Jan Cools, Center for Human Genetics, KULeuven, Leuven for the MOHITO cell line. We thank Dr Phillip Ivanyi and Professor Anke Franzke, Department of Hematology, MHH for providing human T-ALL cell lines. We thank Rena-Mareike Struss, Institute of Experimental Hematology, Hannover Medical School for help with mouse experiments and Anneke Doerrie, Institute of Biochemistry, for assistance in polysome profiling experiments. We thank Axel Schambach, Institute of Experimental Hematology, for providing pRSF91.IRES.dTomato and pRRL.PPT.SF.IRES.EGFP. WPRE vectors. We thank Thomas Neumann and Maike Stahlhut for performing the microarray experiments. We thank the Cell Sorting Unit of Hannover Medical School for performing sorting experiments and Michael Morgan for proofreading the manuscript. This work was supported by the Deutsche Krebshilfe (grants: 108245 and 109800), the Deutsche José Carreras Leukämie-Stiftung (grant: DJCLS F10/06), the Deutsche Forschungsgemeinschaft (grants: Li 1608/2-1 and SFB738) and the DFG-Cluster of Excellence REBIRTH.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Schwarzer, A., Holtmann, H., Brugman, M. et al. Hyperactivation of mTORC1 and mTORC2 by multiple oncogenic events causes addiction to eIF4E-dependent mRNA translation in T-cell leukemia. Oncogene 34, 3593–3604 (2015). https://doi.org/10.1038/onc.2014.290
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2014.290
This article is cited by
-
A phase I study of a dual PI3-kinase/mTOR inhibitor BEZ235 in adult patients with relapsed or refractory acute leukemia
BMC Pharmacology and Toxicology (2020)
-
Repression of oncogenic cap-mediated translation by 4Ei-10 diminishes proliferation, enhances chemosensitivity and alters expression of malignancy-related proteins in mesothelioma
Cancer Chemotherapy and Pharmacology (2020)
-
Inhibition of oncogenic cap-dependent translation by 4EGI-1 reduces growth, enhances chemosensitivity and alters genome-wide translation in non-small cell lung cancer
Cancer Gene Therapy (2019)
-
4EGI-1 represses cap-dependent translation and regulates genome-wide translation in malignant pleural mesothelioma
Investigational New Drugs (2018)
-
NTRK2 activation cooperates with PTEN deficiency in T-ALL through activation of both the PI3K–AKT and JAK–STAT3 pathways
Cell Discovery (2016)