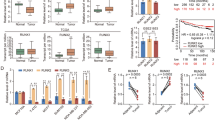
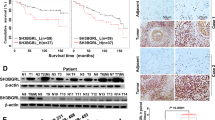
Abstract
Elucidating targets of physiological tumor metastasis suppressors can highlight key signaling pathways leading to invasion and metastasis. To identify downstream targets of the metastasis suppressor Raf-1 kinase inhibitory protein (RKIP/PEBP1), we utilized an integrated approach based upon statistical analysis of tumor gene expression data combined with experimental validation. Previous studies from our laboratory identified the architectural transcription factor and oncogene, high mobility group AT-hook 2 (HMGA2), as a target of inhibition by RKIP. Here we identify two signaling pathways that promote HMGA2-driven metastasis. Using both human breast tumor cells and an MMTV-Wnt mouse breast tumor model, we show that RKIP induces and HMGA2 inhibits expression of miR-200b; miR-200b directly inhibits expression of lysyl oxidase (LOX), leading to decreased invasion. RKIP also inhibits syndecan-2 (SDC2), which is aberrantly expressed in breast cancer, via downregulation of HMGA2; but this mechanism is independent of miR-200. Depletion of SDC2 induces apoptosis and suppresses breast tumor growth and metastasis in mouse xenografts. RKIP, LOX and SDC2 are coordinately regulated and collectively encompass a prognostic signature for metastasis-free survival in ER-negative breast cancer patients. Taken together, our findings reveal two novel signaling pathways targeted by the metastasis suppressor RKIP that regulate remodeling of the extracellular matrix and tumor survival.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Massague J . Sorting out breast-cancer gene signatures. N Engl J Med 2007; 356: 294–297.
Steeg PS . Tumor metastasis: mechanistic insights and clinical challenges. Nat Med 2006; 12: 895–904.
Smith SC, Theodorescu D . Learning therapeutic lessons from metastasis suppressor proteins. Nat Rev Cancer 2009; 9: 253–264.
Fu Z, Smith PC, Zhang L, Rubin MA, Dunn RL, Yao Z et al. Effects of raf kinase inhibitor protein expression on suppression of prostate cancer metastasis. J Natl Cancer Inst 2003; 95: 878–889.
Fu Z, Kitagawa Y, Shen R, Shah R, Mehra R, Rhodes D et al. Metastasis suppressor gene Raf kinase inhibitor protein (RKIP) is a novel prognostic marker in prostate cancer. Prostate 2006; 66: 248–256.
Al-Mulla F, Hagan S, Behbehani AI, Bitar MS, George SS, Going JJ et al. Raf kinase inhibitor protein expression in a survival analysis of colorectal cancer patients. J Clin Oncol 2006; 24: 5672–5679.
Minoo P, Zlobec I, Baker K, Tornillo L, Terracciano L, Jass JR et al. Loss of raf-1 kinase inhibitor protein expression is associated with tumor progression and metastasis in colorectal cancer. Am J Clin Pathol 2007; 127: 820–827.
Yun J, Frankenberger CA, Kuo WL, Boelens MC, Eves EM, Cheng N et al. Signalling pathway for RKIP and Let-7 regulates and predicts metastatic breast cancer. EMBO J 2011; 30: 4500–4514.
Hagan S, Al-Mulla F, Mallon E, Oien K, Ferrier R, Gusterson B et al. Reduction of Raf-1 kinase inhibitor protein expression correlates with breast cancer metastasis. Clin Cancer Res 2005; 11: 7392–7397.
Dangi-Garimella S, Yun J, Eves EM, Newman M, Erkeland SJ, Hammond SM et al. Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving LIN28 and let-7. EMBO J 2009; 28: 347–358.
Xu YF, Yi Y, Qiu SJ, Gao Q, Li YW, Dai CX et al. PEBP1 downregulation is associated to poor prognosis in HCC related to hepatitis B infection. J Hepatol 2010; 53: 872–879.
Yeung K, Seitz T, Li S, Janosch P, McFerran B, Kaiser C et al. Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP. Nature 1999; 401: 173–177.
Corbit KC, Trakul N, Eves EM, Diaz B, Marshall M, Rosner MR . Activation of Raf-1 signaling by protein kinase C through a mechanism involving Raf kinase inhibitory protein. J Biol Chem 2003; 278: 13061–13068.
Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 2008; 10: 593–601.
Park SM, Gaur AB, Lengyel E, Peter ME . The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 2008; 22: 894–907.
Erler JT, Bennewith KL, Nicolau M, Dornhofer N, Kong C, Le QT et al. Lysyl oxidase is essential for hypoxia-induced metastasis. Nature 2006; 440: 1222–1226.
Erler JT, Bennewith KL, Cox TR, Lang G, Bird D, Koong A et al. Hypoxia-induced lysyl oxidase is a critical mediator of bone marrow cell recruitment to form the premetastatic niche. Cancer Cell 2009; 15: 35–44.
Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 2003; 3: 537–549.
Huang da W, Sherman BT, Lempicki RA . Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4: 44–57.
Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ et al. Combinatorial microRNA target predictions. Nat Genet 2005; 37: 495–500.
Lewis BP, Burge CB, Bartel DP . Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005; 120: 15–20.
John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS . Human MicroRNA targets. PLoS Biol 2004; 2: e363.
Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ . miRBase: tools for microRNA genomics. Nucleic Acids Res 2008; 36: D154–D158.
Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E . The role of site accessibility in microRNA target recognition. Nat Genet 2007; 39: 1278–1284.
Efron B, Tibshirani R . On testing the significance of sets of genes. Ann Appl Stat 2007; 1: 107–129.
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D et al. MicroRNA expression profiles classify human cancers. Nature 2005; 435: 834–838.
Csiszar K . Lysyl oxidases: a novel multifunctional amine oxidase family. Prog Nucleic Acid Res Mol Biol 2001; 70: 1–32.
Schuetz CS, Bonin M, Clare SE, Nieselt K, Sotlar K, Walter M et al. Progression-specific genes identified by expression profiling of matched ductal carcinomas in situ and invasive breast tumors, combining laser capture microdissection and oligonucleotide microarray analysis. Cancer Res 2006; 66: 5278–5286.
Peter ME . Let-7 and miR-200 microRNAs: guardians against pluripotency and cancer progression. Cell Cycle 2009; 8: 843–852.
Li Y, VandenBoom TG 2nd, Kong D, Wang Z, Ali S, Philip PA et al. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res 2009; 69: 6704–6712.
Morishita A, Zaidi M, Mitoro A, Sankarasharma D, Szabolcs M, Okada Y et al. HMGA2 is an in vivo driver of tumor metastasis. Cancer Res 2013; 73: 4289–4299.
Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol 2009; 11: 1487–1495.
Lambaerts K, Wilcox-Adelman SA, Zimmermann P . The signaling mechanisms of syndecan heparan sulfate proteoglycans. Curr Opin Cell Biol 2009; 21: 662–669.
Barbareschi M, Maisonneuve P, Aldovini D, Cangi MG, Pecciarini L, Angelo Mauri F et al. High syndecan-1 expression in breast carcinoma is related to an aggressive phenotype and to poorer prognosis. Cancer 2003; 98: 474–483.
Loussouarn D, Campion L, Sagan C, Frenel JS, Dravet F, Classe JM et al. Prognostic impact of syndecan-1 expression in invasive ductal breast carcinomas. Br J Cancer 2008; 98: 1993–1998.
Nikolova V, Koo CY, Ibrahim SA, Wang Z, Spillmann D, Dreier R et al. Differential roles for membrane-bound and soluble syndecan-1 (CD138) in breast cancer progression. Carcinogenesis 2009; 30: 397–407.
De Oliveira T, Abiatari I, Raulefs S, Sauliunaite D, Erkan M, Kong B et al. Syndecan-2 promotes perineural invasion and cooperates with K-ras to induce an invasive pancreatic cancer cell phenotype. Mol Cancer 2012; 11: 19.
Park H, Kim Y, Lim Y, Han I, Oh ES . Syndecan-2 mediates adhesion and proliferation of colon carcinoma cells. J Biol Chem 2002; 277: 29730–29736.
Popovic A, Demirovic A, Spajic B, Stimac G, Kruslin B, Tomas D . Expression and prognostic role of syndecan-2 in prostate cancer. Prostate Cancer Prostatic Dis 2010; 13: 78–82.
Marion A, Dieudonne FX, Patino-Garcia A, Lecanda F, Marie PJ, Modrowski D . Calpain-6 is an endothelin-1 signaling dependent protective factor in chemoresistant osteosarcoma. Int J Cancer 2012; 130: 2514–2525.
Minn AJ, Bevilacqua E, Yun J, Rosner MR . Identification of novel metastasis suppressor signaling pathways for breast cancer. Cell Cycle 2012; 11: 2452–2457.
Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP . Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 2003; 31: e15.
Team RDC R: A language and environment for statistical computing. 2009.
Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 2004; 5: R80.
Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, Holloway A et al. A comparison of background correction methods for two-colour microarrays. Bioinformatics 2007; 23: 2700–2707.
Berger JA, Hautaniemi S, Jarvinen AK, Edgren H, Mitra SK, Astola J . Optimized LOWESS normalization parameter selection for DNA microarray data. BMC Bioinformatics 2004; 5: 194.
Li Z, Lu J, Sun M, Mi S, Zhang H, Luo RT et al. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc Natl Acad Sci USA 2008; 105: 15535–15540.
Jiang X, Huang H, Li Z, Li Y, Wang X, Gurbuxani S et al. Blockade of miR-150 maturation by MLL-Fusion/MYC/LIN-28 is required for MLL-associated leukemia. Cancer Cell 2012; 22: 524–535.
Sun M, Song CX, Huang H, Frankenberger CA, Sankarasharma D, Gomes S et al. HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer growth and metastasis. Proc Natl Acad Sci USA 2013; 110: 9920–9925.
Minn AJ, Gupta GP, Padua D, Bos P, Nguyen DX, Nuyten D et al. Lung metastasis genes couple breast tumor size and metastatic spread. Proc Natl Acad Sci USA 2007; 104: 6740–6745.
Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD et al. Genes that mediate breast cancer metastasis to lung. Nature 2005; 436: 518–524.
Enerly E, Steinfeld I, Kleivi K, Leivonen SK, Aure MR, Russnes HG et al. miRNA-mRNA integrated analysis reveals roles for miRNAs in primary breast tumors. PLoS One 2011; 6: e16915.
Miller LD, Smeds J, George J, Vega VB, Vergara L, Ploner A et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci USA 2005; 102: 13550–13555.
Desmedt C, Piette F, Loi S, Wang Y, Lallemand F, Haibe-Kains B et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res 2007; 13: 3207–3214.
Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 2005; 365: 671–679.
Acknowledgements
This work was supported by NIH grant GM087630 to MRR and DOD Predoctoral Traineeship Award W81XWH-10-1-0396 to MS. We also thank Eva Eves for technical support and helpful discussion.
Author contributions
MS and MRR designed research; MS, SG, PC, CFA and DS performed research; CHC and KKC contributed new reagents or analytic tools; MS analyzed data; MS and MRR wrote the paper, and all the authors helped to revise the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Rights and permissions
About this article
Cite this article
Sun, M., Gomes, S., Chen, P. et al. RKIP and HMGA2 regulate breast tumor survival and metastasis through lysyl oxidase and syndecan-2. Oncogene 33, 3528–3537 (2014). https://doi.org/10.1038/onc.2013.328
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2013.328
Keywords
This article is cited by
-
MiR-200c downregulates HIF-1α and inhibits migration of lung cancer cells
Cellular & Molecular Biology Letters (2019)
-
ACGH detects distinct genomic alterations of primary intrahepatic cholangiocarcinomas and matched lymph node metastases and identifies a poor prognosis subclass
Scientific Reports (2018)
-
A novel long non-coding RNA lnc-GNAT1-1 is low expressed in colorectal cancer and acts as a tumor suppressor through regulating RKIP-NF-κB-Snail circuit
Journal of Experimental & Clinical Cancer Research (2016)
-
Cell surface heparan sulfate proteoglycans control adhesion and invasion of breast carcinoma cells
Molecular Cancer (2015)
-
The biological complexity of RKIP signaling in human cancers
Experimental & Molecular Medicine (2015)