Abstract
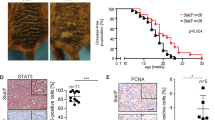
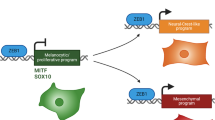
Melanoma is a highly lethal malignancy notorious for its aggressive clinical course and eventual resistance to existing therapies. Currently, we possess a limited understanding of the genetic events driving melanoma progression, and much effort is focused on identifying pro-metastatic aberrations or perturbed signaling networks that constitute new therapeutic targets. In this study, we validate and assess the mechanism by which homeobox transcription factor A1 (HOXA1), a pro-invasion oncogene previously identified in a metastasis screen by our group, contributes to melanoma progression. Transcriptome and pathway profiling analyses of cells expressing HOXA1 reveals upregulation of factors involved in diverse cytokine pathways that include the transforming growth factor beta (TGFβ) signaling axis, which we further demonstrate to be required for HOXA1-mediated cell invasion in melanoma cells. Transcriptome profiling also shows HOXA1’s ability to potently downregulate expression of microphthalmia-associated transcription factor (MITF) and other genes required for melanocyte differentiation, suggesting a mechanism by which HOXA1 expression de-differentiates cells into a pro-invasive cell state concomitant with TGFβ activation. Our analysis of publicly available data sets indicate that the HOXA1-induced gene signature successfully categorizes melanoma specimens based on their metastatic potential and, importantly, is capable of stratifying melanoma patient risk for metastasis based on expression in primary tumors. Together, these validation data and mechanistic insights suggest that patients whose primary tumors express HOXA1 are among a high-risk metastasis subgroup that should be considered for anti-TGFβ therapy in adjuvant settings. Moreover, further analysis of HOXA1 target genes in melanoma may reveal new pathways or targets amenable to therapeutic intervention.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D . Global cancer statistics. CA Cancer J Clin 2011; 61: 69–90.
Valastyan S, Weinberg RA . Tumor metastasis: molecular insights and evolving paradigms. Cell 2011; 147: 275–292.
Bernards R, Weinberg RA . A progression puzzle. Nature 2002; 418: 823.
van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 2002; 347: 1999–2009.
Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 2010; 464: 999–1005.
Breslow A . Thickness, cross-sectional areas and depth of invasion in the prognosis of cutaneous melanoma. Ann Surg 1970; 172: 902–908.
Tuong W, Cheng LS, Armstrong AW . Melanoma: epidemiology, diagnosis, treatment, and outcomes. Dermatol Clin 2012; 30: 113–124.
Gimotty PA, Elder DE, Fraker DL, Botbyl J, Sellers K, Elenitsas R et al. Identification of high-risk patients among those diagnosed with thin cutaneous melanomas. J Clin Oncol 2007; 25: 1129–1134.
Criscione VD, Weinstock MA . Melanoma thickness trends in the United States, 1988-2006. J Invest Dermatol 2010; 130: 793–797.
Scott KL, Nogueira C, Heffernan TP, van Doorn R, Dhakal S, Hanna JA et al. Proinvasion metastasis drivers in early-stage melanoma are oncogenes. Cancer Cell 2011; 20: 92–103.
Chin L, Tam A, Pomerantz J, Wong M, Holash J, Bardeesy N et al. Essential role for oncogenic Ras in tumour maintenance. Nature 1999; 400: 468–472.
Kim M, Gans JD, Nogueira C, Wang A, Paik JH, Feng B et al. Comparative oncogenomics identifies NEDD9 as a melanoma metastasis gene. Cell 2006; 125: 1269–1281.
Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature 2005; 436: 117–122.
Remacle S, Shaw-Jackson C, Matis C, Lampe X, Picard J, Rezsohazy R . Changing homeodomain residues 2 and 3 of Hoxa1 alters its activity in a cell-type and enhancer dependent manner. Nucleic Acids Res 2002; 30: 2663–2668.
Svingen T, Tonissen KF . Hox transcription factors and their elusive mammalian gene targets. Heredity 2006; 97: 88–96.
Kaminska B, Wesolowska A, Danilkiewicz M . TGF beta signalling and its role in tumour pathogenesis. Acta Biochim Pol 2005; 52: 329–337.
Wrana JL, Attisano L, Carcamo J, Zentella A, Doody J, Laiho M et al. TGF beta signals through a heteromeric protein kinase receptor complex. Cell 1992; 71: 1003–1014.
Creighton CJ, Casa A, Lazard Z, Huang S, Tsimelzon A, Hilsenbeck SG et al. Insulin-like growth factor-I activates gene transcription programs strongly associated with poor breast cancer prognosis. J Clin Oncol 2008; 26: 4078–4085.
Hoek KS, Schlegel NC, Brafford P, Sucker A, Ugurel S, Kumar R et al. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res 2006; 19: 290–302.
Jeffs AR, Glover AC, Slobbe LJ, Wang L, He S, Hazlett JA et al. A gene expression signature of invasive potential in metastatic melanoma cells. PLoS ONE 2009; 4: e8461.
Tímár J, Győrffy B, Rásó E . Gene signature of the metastatic potential of cutaneous melanoma: too much for too little? Clin Exp Metastasis 2010; 27: 371–387.
Winnepenninckx V, Lazar V, Michiels S, Dessen P, Stas M, Alonso SR et al. Gene expression profiling of primary cutaneous melanoma and clinical outcome. J Natl Cancer Inst 2006; 98: 472–482.
Shah N, Sukumar S . The Hox genes and their roles in oncogenesis. Nat Rev Cancer 2010; 10: 361–371.
Maeda K, Hamada J, Takahashi Y, Tada M, Yamamoto Y, Sugihara T et al. Altered expressions of HOX genes in human cutaneous malignant melanoma. Int J Cancer 2005; 114: 436–441.
Gavalas A, Trainor P, Ariza-McNaughton L, Krumlauf R . Synergy between Hoxa1 and Hoxb1: the relationship between arch patterning and the generation of cranial neural crest. Development 2001; 128: 3017–3027.
Carreira S, Goodall J, Denat L, Rodriguez M, Nuciforo P, Hoek KS et al. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes Dev 2006; 20: 3426–3439.
Lekmine F, Chang CK, Sethakorn N, Das Gupta TK, Salti GI . Role of microphthalmia transcription factor (Mitf) in melanoma differentiation. Biochem Biophys Res Commun 2007; 354: 830–835.
Hoek KS, Goding CR . Cancer stem cells versus phenotype-switching in melanoma. Pigment Cell Melanoma Res 2010; 23: 746–759.
Hoek KS, Eichhoff OM, Schlegel NC, Dobbeling U, Kobert N, Schaerer L et al. In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Res 2008; 68: 650–656.
Nishimura EK, Suzuki M, Igras V, Du J, Lonning S, Miyachi Y et al. Key roles for transforming growth factor β in melanocyte stem cell maintenance. Stem Cell 2010; 6: 130–140.
Pinner S, Jordan P, Sharrock K, Bazley L, Collinson L, Marais R et al. Intravital imaging reveals transient changes in pigment production and Brn2 expression during metastatic melanoma dissemination. Cancer Res 2009; 69: 7969–7977.
Mohammad KS, Javelaud D, Fournier PGJ, Niewolna M, Mckenna CR, Peng XH et al. TGF- -RI kinase inhibitor SD-208 reduces the development and progression of melanoma bone metastases. Cancer Res 2011; 71: 175–184.
Meerbrey KL, Hu G, Kessler JD, Roarty K, Li MZ, Fang JE et al. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc Natl Acad Sci USA 2011; 108: 3665–3670.
He W, Dorn DC, Erdjument-Bromage H, Tempst P, Moore MAS, Massagué J . Hematopoiesis controlled by distinct TIF1gamma and Smad4 branches of the TGFbeta pathway. Cell 2006; 125: 929–941.
Storey JD, Tibshirani R . Statistical significance for genomewide studies. Proc Natl Acad Sci USA 2003; 100: 9440–9445.
Acknowledgements
KLS was supported by the American Cancer Society, and this work was supported by departmental seed funds to KLS from Baylor College of Medicine. This work was also supported by grants from the NIH (RO1 CA93947, U01 CA84313) to LC.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Wardwell-Ozgo, J., Dogruluk, T., Gifford, A. et al. HOXA1 drives melanoma tumor growth and metastasis and elicits an invasion gene expression signature that prognosticates clinical outcome. Oncogene 33, 1017–1026 (2014). https://doi.org/10.1038/onc.2013.30
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2013.30
Keywords
This article is cited by
-
Role of PLK1/NUMB/NOTCH in epithelial-mesenchymal transition in human melanoma
npj Precision Oncology (2024)
-
CircTRRAP (hsa_circ_0081234) participates in prostate cancer progression and glycolysis by HOXA1 via functioning as a miR-515-5p sponge
Applied Biological Chemistry (2022)
-
Molecular implications of HOX genes targeting multiple signaling pathways in cancer
Cell Biology and Toxicology (2022)
-
Protein expression of prognostic genes in primary melanoma and benign nevi
Journal of Cancer Research and Clinical Oncology (2022)
-
Phenotype plasticity as enabler of melanoma progression and therapy resistance
Nature Reviews Cancer (2019)